ARGONAUTE2 cooperates with SWI/SNF complex to determine nucleosome occupancy at human Transcription Start Sites
- PMID: 25605800
- PMCID: PMC4330357
- DOI: 10.1093/nar/gku1387
ARGONAUTE2 cooperates with SWI/SNF complex to determine nucleosome occupancy at human Transcription Start Sites
Abstract
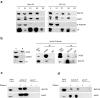
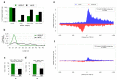
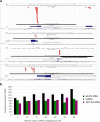
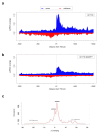
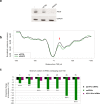
Argonaute (AGO) proteins have a well-established role in post-transcriptional regulation of gene expression as key component of the RNA silencing pathways. Recent evidence involves AGO proteins in mammalian nuclear processes such as transcription and splicing, though the mechanistic aspects of AGO nuclear functions remain largely elusive. Here, by SILAC-based interaction proteomics, we identify the chromatin-remodelling complex SWI/SNF as a novel AGO2 interactor in human cells. Moreover, we show that nuclear AGO2 is loaded with a novel class of Dicer-dependent short RNAs (sRNAs), that we called swiRNAs, which map nearby the Transcription Start Sites (TSSs) bound by SWI/SNF. The knock-down of AGO2 decreases nucleosome occupancy at the first nucleosome located downstream of TSSs in a swiRNA-dependent manner. Our findings indicate that in human cells AGO2 binds SWI/SNF and a novel class of sRNAs to establish nucleosome occupancy on target TSSs.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex.Science. 1996 Jul 26;273(5274):513-6. doi: 10.1126/science.273.5274.513. Science. 1996. PMID: 8662543
-
Human SWI/SNF drives sequence-directed repositioning of nucleosomes on C-myc promoter DNA minicircles.Biochemistry. 2007 Oct 9;46(40):11377-88. doi: 10.1021/bi7008823. Epub 2007 Sep 18. Biochemistry. 2007. PMID: 17877373 Free PMC article.
-
SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast.Genes Dev. 2018 May 1;32(9-10):695-710. doi: 10.1101/gad.312850.118. Epub 2018 May 21. Genes Dev. 2018. PMID: 29785963 Free PMC article.
-
Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence.Biochem Cell Biol. 2007 Aug;85(4):419-25. doi: 10.1139/O07-070. Biochem Cell Biol. 2007. PMID: 17713577 Review.
-
BAFfling pathologies: Alterations of BAF complexes in cancer.Cancer Lett. 2018 Apr 10;419:266-279. doi: 10.1016/j.canlet.2018.01.046. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29374542 Review.
Cited by
-
Argonaute 2 Binds Directly to tRNA Genes and Promotes Gene Repression in cis.Mol Cell Biol. 2015 Jul;35(13):2278-94. doi: 10.1128/MCB.00076-15. Mol Cell Biol. 2015. PMID: 25918241 Free PMC article.
-
Downstream Antisense Transcription Predicts Genomic Features That Define the Specific Chromatin Environment at Mammalian Promoters.PLoS Genet. 2016 Aug 3;12(8):e1006224. doi: 10.1371/journal.pgen.1006224. eCollection 2016 Aug. PLoS Genet. 2016. PMID: 27487356 Free PMC article.
-
Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer.BMC Cancer. 2019 Jul 19;19(1):712. doi: 10.1186/s12885-019-5884-x. BMC Cancer. 2019. PMID: 31324173 Free PMC article.
-
The nuclear receptor ERβ engages AGO2 in regulation of gene transcription, RNA splicing and RISC loading.Genome Biol. 2017 Oct 6;18(1):189. doi: 10.1186/s13059-017-1321-0. Genome Biol. 2017. PMID: 29017520 Free PMC article.
-
RNA matchmaking in chromatin regulation.Biochem Soc Trans. 2020 Dec 18;48(6):2467-2481. doi: 10.1042/BST20191225. Biochem Soc Trans. 2020. PMID: 33245317 Free PMC article. Review.
References
-
- Romano N., Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol. Microbiol. 1992;6:3343–3353. - PubMed
-
- Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. - PubMed
-
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

