Centrosomal nucleolin is required for microtubule network organization
- PMID: 25590348
- PMCID: PMC4614815
- DOI: 10.1080/15384101.2014.1000197
Centrosomal nucleolin is required for microtubule network organization
Abstract
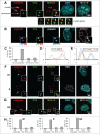
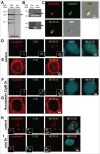
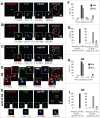
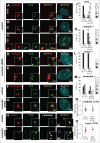
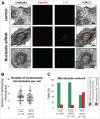
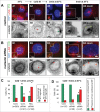
Nucleolin is a pleiotropic protein involved in a variety of cellular processes. Although multipolar spindle formation has been observed after nucleolin depletion, the roles of nucleolin in centrosome regulation and functions have not been addressed. Here we report using immunofluorescence and biochemically purified centrosomes that nucleolin co-localized only with one of the centrioles during interphase which was further identified as the mature centriole. Upon nucleolin depletion, cells exhibited an amplification of immature centriole markers surrounded by irregular pericentrin staining; these structures were exempt from maturation markers and unable to nucleate microtubules. Furthermore, the microtubule network was disorganized in these cells, exhibiting frequent non-centrosomal microtubules. At the mature centriole a reduced kinetics in the centrosomal microtubule nucleation phase was observed in live silenced cells, as well as a perturbation of microtubule anchoring. Immunoprecipitation experiments showed that nucleolin belongs to protein complexes containing 2 key centrosomal proteins, γ-tubulin and ninein, involved in microtubule nucleation and anchoring steps. Altogether, our study uncovered a new role for nucleolin in restricting microtubule nucleation and anchoring at centrosomes in interphase cells.
Keywords: centrosome; interphase; mature centriole; microtubules; nucleolin.
Figures


Similar articles
-
Expression of Nucleolin Affects Microtubule Dynamics.PLoS One. 2016 Jun 16;11(6):e0157534. doi: 10.1371/journal.pone.0157534. eCollection 2016. PLoS One. 2016. PMID: 27309529 Free PMC article.
-
Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly.Mol Biol Cell. 2006 Feb;17(2):680-9. doi: 10.1091/mbc.e05-04-0360. Epub 2005 Nov 16. Mol Biol Cell. 2006. PMID: 16291865 Free PMC article.
-
Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function.J Cell Sci. 2005 Apr 15;118(Pt 8):1565-75. doi: 10.1242/jcs.02302. Epub 2005 Mar 22. J Cell Sci. 2005. PMID: 15784680
-
Centrosome function: sometimes less is more.Traffic. 2009 May;10(5):472-81. doi: 10.1111/j.1600-0854.2009.00880.x. Epub 2009 Jan 24. Traffic. 2009. PMID: 19192251 Review.
-
Regulation of microtubule nucleation mediated by γ-tubulin complexes.Protoplasma. 2017 May;254(3):1187-1199. doi: 10.1007/s00709-016-1070-z. Epub 2017 Jan 10. Protoplasma. 2017. PMID: 28074286 Review.
Cited by
-
Cancer Stem Cells and Nucleolin as Drivers of Carcinogenesis.Pharmaceuticals (Basel). 2021 Jan 13;14(1):60. doi: 10.3390/ph14010060. Pharmaceuticals (Basel). 2021. PMID: 33451077 Free PMC article. Review.
-
Nucleolin is required for multiple centrosome-associated functions in early vertebrate mitosis.Chromosoma. 2023 Nov;132(4):305-315. doi: 10.1007/s00412-023-00808-4. Epub 2023 Aug 24. Chromosoma. 2023. PMID: 37615728
-
Integrated analysis of mRNA and miRNA expression in HeLa cells expressing low levels of Nucleolin.Sci Rep. 2017 Aug 21;7(1):9017. doi: 10.1038/s41598-017-09353-4. Sci Rep. 2017. PMID: 28827664 Free PMC article.
-
Innovative particle standards and long-lived imaging for 2D and 3D dSTORM.Sci Rep. 2019 Nov 29;9(1):17967. doi: 10.1038/s41598-019-53528-0. Sci Rep. 2019. PMID: 31784555 Free PMC article.
-
Nucleolin: a cell portal for viruses, bacteria, and toxins.Cell Mol Life Sci. 2022 May 3;79(5):271. doi: 10.1007/s00018-022-04300-7. Cell Mol Life Sci. 2022. PMID: 35503380 Free PMC article. Review.
References
-
- Bornens M. The centrosome in cells and organisms. Science 2012; 335:422-6; PMID:22282802; http://dx.doi.org/10.1126/science.1209037 - DOI - PubMed
-
- Gonczy P. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol 2012; 13:425-35; PMID:22691849; http://dx.doi.org/10.1038/nrm3373 - DOI - PubMed
-
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol 2007; 17:215-21; PMID:17383880; http://dx.doi.org/10.1016/j.tcb.2007.03.003 - DOI - PubMed
-
- Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 2011; 13:1154-60; PMID:21968988; http://dx.doi.org/10.1038/ncb2345 - DOI - PMC - PubMed
-
- Meraldi P, Nigg EA. The centrosome cycle. FEBS Lett 2002; 521:9-13; PMID:12067716; http://dx.doi.org/10.1016/S0014-5793(02)02865-X - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
