Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication
- PMID: 25589642
- PMCID: PMC4337542
- DOI: 10.1128/JVI.02677-14
Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication
Abstract
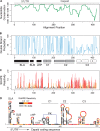
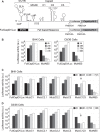
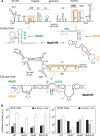
The dengue virus genome is a dynamic molecule that adopts different conformations in the infected cell. Here, using RNA folding predictions, chemical probing analysis, RNA binding assays, and functional studies, we identified new cis-acting elements present in the capsid coding sequence that facilitate cyclization of the viral RNA by hybridization with a sequence involved in a local dumbbell structure at the viral 3' untranslated region (UTR). The identified interaction differentially enhances viral replication in mosquito and mammalian cells.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization.J Virol. 2013 Jun;87(12):6804-18. doi: 10.1128/JVI.00243-13. Epub 2013 Apr 10. J Virol. 2013. PMID: 23576500 Free PMC article.
-
RNA Structure Duplication in the Dengue Virus 3' UTR: Redundancy or Host Specificity?mBio. 2019 Jan 8;10(1):e02506-18. doi: 10.1128/mBio.02506-18. mBio. 2019. PMID: 30622191 Free PMC article.
-
Uncoupling cis-Acting RNA elements from coding sequences revealed a requirement of the N-terminal region of dengue virus capsid protein in virus particle formation.J Virol. 2012 Jan;86(2):1046-58. doi: 10.1128/JVI.05431-11. Epub 2011 Nov 9. J Virol. 2012. PMID: 22072762 Free PMC article.
-
Dynamic RNA structures in the dengue virus genome.RNA Biol. 2011 Mar-Apr;8(2):249-57. doi: 10.4161/rna.8.2.14992. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21593583 Review.
-
Functional RNA elements in the dengue virus genome.Viruses. 2011 Sep;3(9):1739-56. doi: 10.3390/v3091739. Epub 2011 Sep 15. Viruses. 2011. PMID: 21994804 Free PMC article. Review.
Cited by
-
The tale of two flaviviruses: subversion of host pathways by RNA shapes in dengue and hepatitis C viral RNA genomes.Curr Opin Microbiol. 2021 Feb;59:79-85. doi: 10.1016/j.mib.2020.08.007. Epub 2020 Oct 16. Curr Opin Microbiol. 2021. PMID: 33070015 Free PMC article. Review.
-
Evolutionary traits of Tick-borne encephalitis virus: Pervasive non-coding RNA structure conservation and molecular epidemiology.Virus Evol. 2022 Jun 11;8(1):veac051. doi: 10.1093/ve/veac051. eCollection 2022. Virus Evol. 2022. PMID: 35822110 Free PMC article.
-
Effects of Refolding on Large-Scale RNA Structure.Biochemistry. 2019 Jul 16;58(28):3069-3077. doi: 10.1021/acs.biochem.8b01219. Epub 2019 Jul 3. Biochemistry. 2019. PMID: 31268687 Free PMC article.
-
Flavors of Flaviviral RNA Structure: towards an Integrated View of RNA Function from Translation through Encapsidation.Bioessays. 2019 Aug;41(8):e1900003. doi: 10.1002/bies.201900003. Epub 2019 Jun 18. Bioessays. 2019. PMID: 31210384 Free PMC article. Review.
-
Genomic variations in the 3'-termini of Rice stripe virus in the rotation between vector insect and host plant.New Phytol. 2018 Aug;219(3):1085-1096. doi: 10.1111/nph.15246. Epub 2018 Jun 8. New Phytol. 2018. PMID: 29882354 Free PMC article.
References
-
- Gamarnik AV. 2010. Role of the dengue virus 5′ and 3′ untranslated regions in viral replication, p 55–78 InHanley KA, Weaver SC (ed), Frontiers in dengue virus research. Caister Academic Press, Wymondham, Norfolk, United Kingdom.
-
- Lindenbach BD, Thiel HJ, Rice CD. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 InKnipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Alcaraz-Estrada SL, Yocupicio-Monroy M, del Angel RM. 2010. Insights into dengue virus genome replication. Future Virol 5:575–592. doi:10.2217/fvl.10.49. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

