Membrane trafficking in the yeast Saccharomyces cerevisiae model
- PMID: 25584613
- PMCID: PMC4307317
- DOI: 10.3390/ijms16011509
Membrane trafficking in the yeast Saccharomyces cerevisiae model
Abstract
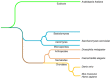
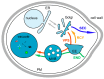
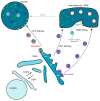
The yeast Saccharomyces cerevisiae is one of the best characterized eukaryotic models. The secretory pathway was the first trafficking pathway clearly understood mainly thanks to the work done in the laboratory of Randy Schekman in the 1980s. They have isolated yeast sec mutants unable to secrete an extracellular enzyme and these SEC genes were identified as encoding key effectors of the secretory machinery. For this work, the 2013 Nobel Prize in Physiology and Medicine has been awarded to Randy Schekman; the prize is shared with James Rothman and Thomas Südhof. Here, we present the different trafficking pathways of yeast S. cerevisiae. At the Golgi apparatus newly synthesized proteins are sorted between those transported to the plasma membrane (PM), or the external medium, via the exocytosis or secretory pathway (SEC), and those targeted to the vacuole either through endosomes (vacuolar protein sorting or VPS pathway) or directly (alkaline phosphatase or ALP pathway). Plasma membrane proteins can be internalized by endocytosis (END) and transported to endosomes where they are sorted between those targeted for vacuolar degradation and those redirected to the Golgi (recycling or RCY pathway). Studies in yeast S. cerevisiae allowed the identification of most of the known effectors, protein complexes, and trafficking pathways in eukaryotic cells, and most of them are conserved among eukaryotes.
Figures

Similar articles
-
Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae.Biochim Biophys Acta. 2005 Jul 10;1744(3):438-54. doi: 10.1016/j.bbamcr.2005.04.004. Biochim Biophys Acta. 2005. PMID: 15913810 Review.
-
Antagonistic roles of ESCRT and Vps class C/HOPS complexes in the recycling of yeast membrane proteins.Mol Biol Cell. 2004 Sep;15(9):4203-14. doi: 10.1091/mbc.e04-05-0420. Epub 2004 Jun 23. Mol Biol Cell. 2004. PMID: 15215319 Free PMC article.
-
Retrograde trafficking and plasma membrane recycling pathways of the budding yeast Saccharomyces cerevisiae.Traffic. 2020 Jan;21(1):45-59. doi: 10.1111/tra.12693. Epub 2019 Oct 1. Traffic. 2020. PMID: 31471931 Review.
-
Yeast membrane lipid imbalance leads to trafficking defects toward the Golgi.Cell Biol Int. 2018 Jul;42(7):890-902. doi: 10.1002/cbin.10956. Epub 2018 Apr 19. Cell Biol Int. 2018. PMID: 29500884
-
Endosomal trafficking of yeast membrane proteins.Biochem Soc Trans. 2018 Dec 17;46(6):1551-1558. doi: 10.1042/BST20180258. Epub 2018 Oct 31. Biochem Soc Trans. 2018. PMID: 30381337 Free PMC article. Review.
Cited by
-
Opsin 1 and Opsin 2 of the Corn Smut Fungus Ustilago maydis Are Green Light-Driven Proton Pumps.Front Microbiol. 2019 Apr 10;10:735. doi: 10.3389/fmicb.2019.00735. eCollection 2019. Front Microbiol. 2019. PMID: 31024506 Free PMC article.
-
Defects in intracellular trafficking of fungal cell wall synthases lead to aberrant host immune recognition.PLoS Pathog. 2018 Jun 4;14(6):e1007126. doi: 10.1371/journal.ppat.1007126. eCollection 2018 Jun. PLoS Pathog. 2018. PMID: 29864141 Free PMC article.
-
Identification of the endocytic sorting signal recognized by the Art1-Rsp5 ubiquitin ligase complex.Mol Biol Cell. 2016 Dec 15;27(25):4043-4054. doi: 10.1091/mbc.E16-08-0570. Epub 2016 Oct 19. Mol Biol Cell. 2016. PMID: 27798240 Free PMC article.
-
Innovation and constraint leading to complex multicellularity in the Ascomycota.Nat Commun. 2017 Feb 8;8:14444. doi: 10.1038/ncomms14444. Nat Commun. 2017. PMID: 28176784 Free PMC article.
-
FgVAC1 is an Essential Gene Required for Golgi-to-Vacuole Transport and Fungal Development in Fusarium graminearum.J Microbiol. 2024 Aug;62(8):649-660. doi: 10.1007/s12275-024-00160-x. Epub 2024 Jul 30. J Microbiol. 2024. PMID: 39080148 Free PMC article.
References
-
- Amberg D.C., Burke D.J., Strathern J.N. Methods in Yeast Genetics. 2005 ed. Cold Spring Harbor Laboratory Press; New York, NY, USA: 2005.
-
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. Life with 6000 genes. Science. 1996;274:563–547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

