Guidelines for monitoring autophagy in Caenorhabditis elegans
- PMID: 25569839
- PMCID: PMC4502811
- DOI: 10.1080/15548627.2014.1003478
Guidelines for monitoring autophagy in Caenorhabditis elegans
Abstract
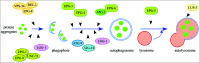
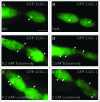
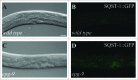
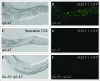
The cellular recycling process of autophagy has been extensively characterized with standard assays in yeast and mammalian cell lines. In multicellular organisms, numerous external and internal factors differentially affect autophagy activity in specific cell types throughout the stages of organismal ontogeny, adding complexity to the analysis of autophagy in these metazoans. Here we summarize currently available assays for monitoring the autophagic process in the nematode C. elegans. A combination of measuring levels of the lipidated Atg8 ortholog LGG-1, degradation of well-characterized autophagic substrates such as germline P granule components and the SQSTM1/p62 ortholog SQST-1, expression of autophagic genes and electron microscopy analysis of autophagic structures are presently the most informative, yet steady-state, approaches available to assess autophagy levels in C. elegans. We also review how altered autophagy activity affects a variety of biological processes in C. elegans such as L1 survival under starvation conditions, dauer formation, aging, and cell death, as well as neuronal cell specification. Taken together, C. elegans is emerging as a powerful model organism to monitor autophagy while evaluating important physiological roles for autophagy in key developmental events as well as during adulthood.
Keywords: ASEL, ASE left; ASER, ASE right; ATG, autophagy-related; C. elegans; ER, endoplasmic reticulum; GFP, green fluorescent protein; LC3; MO, membranous organelle; PGL, P-granule abnormality; RER, rough endoplasmic reticulum; SQST, SeQueSTosome related protein; SQSTM1; TEM, transmission electron microscopy; autophagy; development; epg, ectopic PGL granules; lgg-1, LC3, GABARAP and GATE-16 family.
Figures




Similar articles
-
The tissue- and developmental stage-specific involvement of autophagy genes in aggrephagy.Autophagy. 2020 Apr;16(4):589-599. doi: 10.1080/15548627.2019.1632121. Epub 2019 Jun 26. Autophagy. 2020. PMID: 31204564 Free PMC article.
-
Western blot analysis of the autophagosomal membrane protein LGG-1/LC3 in Caenorhabditis elegans.Methods Enzymol. 2019;619:319-336. doi: 10.1016/bs.mie.2018.12.034. Epub 2019 Feb 4. Methods Enzymol. 2019. PMID: 30910027
-
Selective autophagic degradation of maternally-loaded germline P granule components in somatic cells during C. elegans embryogenesis.Autophagy. 2009 Jul;5(5):717-9. doi: 10.4161/auto.5.5.8552. Epub 2009 Jul 26. Autophagy. 2009. PMID: 19372764
-
Autophagy and innate immunity: Insights from invertebrate model organisms.Autophagy. 2018;14(2):233-242. doi: 10.1080/15548627.2017.1389824. Epub 2018 Feb 17. Autophagy. 2018. PMID: 29130360 Free PMC article. Review.
-
Aggrephagy: lessons from C. elegans.Biochem J. 2013 Jun 15;452(3):381-90. doi: 10.1042/BJ20121721. Biochem J. 2013. PMID: 23725457 Review.
Cited by
-
A fluorescent reporter for rapid assessment of autophagic flux reveals unique autophagy signatures during C. elegans post-embryonic development and identifies compounds that modulate autophagy.Autophagy Rep. 2024;3(1):2371736. doi: 10.1080/27694127.2024.2371736. Epub 2024 Jul 11. Autophagy Rep. 2024. PMID: 39070663
-
Glucosamine Extends the Lifespan of Caenorhabditis elegans via Autophagy Induction.J Appl Glycosci (1999). 2018 Aug 20;65(3):37-43. doi: 10.5458/jag.jag.JAG-2018_002. eCollection 2018. J Appl Glycosci (1999). 2018. PMID: 34354511 Free PMC article.
-
Cysteine Leukotriene Receptor Antagonist-Montelukast Effects on Diabetic Retinal Microvascular Endothelial Cells Curtail Autophagy.Invest Ophthalmol Vis Sci. 2024 Nov 4;65(13):15. doi: 10.1167/iovs.65.13.15. Invest Ophthalmol Vis Sci. 2024. PMID: 39504050 Free PMC article.
-
Methods to Determine the Role of Autophagy Proteins in C. elegans Aging.Methods Mol Biol. 2019;1880:561-586. doi: 10.1007/978-1-4939-8873-0_37. Methods Mol Biol. 2019. PMID: 30610723 Free PMC article.
-
The autophagy receptor p62/SQST-1 promotes proteostasis and longevity in C. elegans by inducing autophagy.Nat Commun. 2019 Dec 11;10(1):5648. doi: 10.1038/s41467-019-13540-4. Nat Commun. 2019. PMID: 31827090 Free PMC article.
References
-
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458-67; PMID:19491929; http://dx.doi.org/10.1038/nrm2708 - DOI - PubMed
-
- Feng YC, He D, Yao ZY, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/10.1038/cr.2013.168 - DOI - PMC - PubMed
-
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27-42; PMID:18191218; http://dx.doi.org/10.1016/j.cell.2007.12.018 - DOI - PMC - PubMed
-
- Tian Y, Li ZP, Hu WQ, Ren HY, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, et al. . C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 2010; 141:1042-55; PMID:20550938; http://dx.doi.org/10.1016/j.cell.2010.04.034 - DOI - PubMed
-
- Lu Q, Yang PG, Huang XX, Hu WQ, Guo B, Wu F, Lin L, Kovács AL, Yu L, Zhang H. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 2011; 21:343-57; PMID:21802374; http://dx.doi.org/10.1016/j.devcel.2011.06.024 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
