Genomic perspectives of transcriptional regulation in forebrain development
- PMID: 25569346
- PMCID: PMC4438709
- DOI: 10.1016/j.neuron.2014.11.011
Genomic perspectives of transcriptional regulation in forebrain development
Abstract
The forebrain is the seat of higher-order brain functions, and many human neuropsychiatric disorders are due to genetic defects affecting forebrain development, making it imperative to understand the underlying genetic circuitry. Recent progress now makes it possible to begin fully elucidating the genomic regulatory mechanisms that control forebrain gene expression. Herein, we discuss the current knowledge of how transcription factors drive gene expression programs through their interactions with cis-acting genomic elements, such as enhancers; how analyses of chromatin and DNA modifications provide insights into gene expression states; and how these approaches yield insights into the evolution of the human brain.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
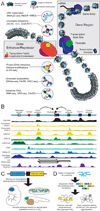


Similar articles
-
Conserved Noncoding Sequences Regulate lhx5 Expression in the Zebrafish Forebrain.PLoS One. 2015 Jul 6;10(7):e0132525. doi: 10.1371/journal.pone.0132525. eCollection 2015. PLoS One. 2015. PMID: 26147098 Free PMC article.
-
Genomic code for Sox2 binding uncovers its regulatory role in Six3 activation in the forebrain.Dev Biol. 2013 Sep 15;381(2):491-501. doi: 10.1016/j.ydbio.2013.06.016. Epub 2013 Jun 19. Dev Biol. 2013. PMID: 23792023
-
Transcriptional enhancers: from properties to genome-wide predictions.Nat Rev Genet. 2014 Apr;15(4):272-86. doi: 10.1038/nrg3682. Epub 2014 Mar 11. Nat Rev Genet. 2014. PMID: 24614317 Review.
-
Identification of Arx transcriptional targets in the developing basal forebrain.Hum Mol Genet. 2008 Dec 1;17(23):3740-60. doi: 10.1093/hmg/ddn271. Epub 2008 Sep 16. Hum Mol Genet. 2008. PMID: 18799476 Free PMC article.
-
Deciphering the transcriptional cis-regulatory code.Trends Genet. 2013 Jan;29(1):11-22. doi: 10.1016/j.tig.2012.09.007. Epub 2012 Oct 23. Trends Genet. 2013. PMID: 23102583 Review.
Cited by
-
Sox2-Dependent 3D Chromatin Interactomes in Transcription, Neural Stem Cell Proliferation and Neurodevelopmental Diseases.J Exp Neurosci. 2019 Aug 7;13:1179069519868224. doi: 10.1177/1179069519868224. eCollection 2019. J Exp Neurosci. 2019. PMID: 31431802 Free PMC article.
-
Traumatic brain injury recapitulates developmental changes of axons.Prog Neurobiol. 2022 Oct;217:102332. doi: 10.1016/j.pneurobio.2022.102332. Epub 2022 Jul 21. Prog Neurobiol. 2022. PMID: 35870679 Free PMC article.
-
Extended intergenic DNA contributes to neuron-specific expression of neighboring genes in the mammalian nervous system.Nat Commun. 2022 May 18;13(1):2733. doi: 10.1038/s41467-022-30192-z. Nat Commun. 2022. PMID: 35585070 Free PMC article.
-
Sp9 Regulates Medial Ganglionic Eminence-Derived Cortical Interneuron Development.Cereb Cortex. 2019 Jun 1;29(6):2653-2667. doi: 10.1093/cercor/bhy133. Cereb Cortex. 2019. PMID: 29878134 Free PMC article.
-
ARNT2 Tunes Activity-Dependent Gene Expression through NCoR2-Mediated Repression and NPAS4-Mediated Activation.Neuron. 2019 Apr 17;102(2):390-406.e9. doi: 10.1016/j.neuron.2019.02.007. Epub 2019 Mar 4. Neuron. 2019. PMID: 30846309 Free PMC article.
References
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. - PubMed
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b;19:27–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HG003988/HG/NHGRI NIH HHS/United States
- T32 GM007618/GM/NIGMS NIH HHS/United States
- R01NS34661/NS/NINDS NIH HHS/United States
- R25 MH077823/MH/NIMH NIH HHS/United States
- T32 GMO7618/PHS HHS/United States
- R01 NS034661/NS/NINDS NIH HHS/United States
- R01NS062859A/NS/NINDS NIH HHS/United States
- F32 GM105202/GM/NIGMS NIH HHS/United States
- R01 MH081880/MH/NIMH NIH HHS/United States
- GM105202/GM/NIGMS NIH HHS/United States
- R01MH081880/MH/NIMH NIH HHS/United States
- R37MH049428/MH/NIMH NIH HHS/United States
- R01HG003988/HG/NHGRI NIH HHS/United States
- R01 NS062859/NS/NINDS NIH HHS/United States
- R37 MH049428/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

