A discrete transition zone organizes the topological and regulatory autonomy of the adjacent tfap2c and bmp7 genes
- PMID: 25569170
- PMCID: PMC4288730
- DOI: 10.1371/journal.pgen.1004897
A discrete transition zone organizes the topological and regulatory autonomy of the adjacent tfap2c and bmp7 genes
Abstract
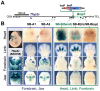
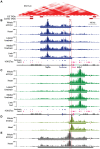
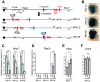
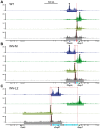
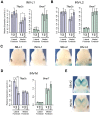
Despite the well-documented role of remote enhancers in controlling developmental gene expression, the mechanisms that allocate enhancers to genes are poorly characterized. Here, we investigate the cis-regulatory organization of the locus containing the Tfap2c and Bmp7 genes in vivo, using a series of engineered chromosomal rearrangements. While these genes lie adjacent to one another, we demonstrate that they are independently regulated by distinct sets of enhancers, which in turn define non-overlapping regulatory domains. Chromosome conformation capture experiments reveal a corresponding partition of the locus in two distinct structural entities, demarcated by a discrete transition zone. The impact of engineered chromosomal rearrangements on the topology of the locus and the resultant gene expression changes indicate that this transition zone functionally organizes the structural partition of the locus, thereby defining enhancer-target gene allocation. This partition is, however, not absolute: we show that it allows competing interactions across it that may be non-productive for the competing gene, but modulate expression of the competed one. Altogether, these data highlight the prime role of the topological organization of the genome in long-distance regulation of gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Comparative cis-regulatory analyses identify new elements of the mouse Hoxc8 early enhancer.J Exp Zool B Mol Dev Evol. 2004 Sep 15;302(5):436-45. doi: 10.1002/jez.b.21009. J Exp Zool B Mol Dev Evol. 2004. PMID: 15384168
-
An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape.Dev Cell. 2013 Mar 11;24(5):530-42. doi: 10.1016/j.devcel.2013.01.025. Epub 2013 Feb 28. Dev Cell. 2013. PMID: 23453598
-
Quail myoD is regulated by a complex array of cis-acting control sequences.Dev Biol. 1995 Jul;170(1):21-38. doi: 10.1006/dbio.1995.1192. Dev Biol. 1995. PMID: 7601311
-
Developmental enhancers and chromosome topology.Science. 2018 Sep 28;361(6409):1341-1345. doi: 10.1126/science.aau0320. Science. 2018. PMID: 30262496 Free PMC article. Review.
-
The architecture of gene expression: integrating dispersed cis-regulatory modules into coherent regulatory domains.Curr Opin Genet Dev. 2014 Aug;27:74-82. doi: 10.1016/j.gde.2014.03.014. Epub 2014 Jun 5. Curr Opin Genet Dev. 2014. PMID: 24907448 Review.
Cited by
-
Minor Loops in Major Folds: Enhancer-Promoter Looping, Chromatin Restructuring, and Their Association with Transcriptional Regulation and Disease.PLoS Genet. 2015 Dec 3;11(12):e1005640. doi: 10.1371/journal.pgen.1005640. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26632825 Free PMC article. Review.
-
Topologically associating domain boundaries are required for normal genome function.Commun Biol. 2023 Apr 20;6(1):435. doi: 10.1038/s42003-023-04819-w. Commun Biol. 2023. PMID: 37081156 Free PMC article.
-
Functional Characterization of a Dual Enhancer/Promoter Regulatory Element Leading Human CD69 Expression.Front Genet. 2020 Oct 27;11:552949. doi: 10.3389/fgene.2020.552949. eCollection 2020. Front Genet. 2020. PMID: 33193627 Free PMC article.
-
Enhancer-promoter interactions can form independently of genomic distance and be functional across TAD boundaries.Nucleic Acids Res. 2024 Feb 28;52(4):1702-1719. doi: 10.1093/nar/gkad1183. Nucleic Acids Res. 2024. PMID: 38084924 Free PMC article.
-
Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors.Trends Genet. 2016 Dec;32(12):801-814. doi: 10.1016/j.tig.2016.10.003. Epub 2016 Nov 2. Trends Genet. 2016. PMID: 27816209 Free PMC article. Review.
References
-
- De Gobbi M (2006) A Regulatory SNP Causes a Human Genetic Disease by Creating a New Transcriptional Promoter. Science 312: 1215–1217. - PubMed
-
- Peichel CL, Prabhakaran B, Vogt TF (1997) The mouse Ulnaless mutation deregulates posterior HoxD gene expression and alters appendicular patterning. Development 124: 3481–3492. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

