The role of cellular adhesion molecules in virus attachment and entry
- PMID: 25533093
- PMCID: PMC4275905
- DOI: 10.1098/rstb.2014.0035
The role of cellular adhesion molecules in virus attachment and entry
Abstract
As obligate intracellular parasites, viruses must traverse the host-cell plasma membrane to initiate infection. This presents a formidable barrier, which they have evolved diverse strategies to overcome. Common to all entry pathways, however, is a mechanism of specific attachment to cell-surface macromolecules or 'receptors'. Receptor usage frequently defines viral tropism, and consequently, the evolutionary changes in receptor specificity can lead to emergence of new strains exhibiting altered pathogenicity or host range. Several classes of molecules are exploited as receptors by diverse groups of viruses, including, for example, sialic acid moieties and integrins. In particular, many cell-adhesion molecules that belong to the immunoglobulin-like superfamily of proteins (IgSF CAMs) have been identified as viral receptors. Structural analysis of the interactions between viruses and IgSF CAM receptors has not shown binding to specific features, implying that the Ig-like fold may not be key. Both proteinaceous and enveloped viruses exploit these proteins, however, suggesting convergent evolution of this trait. Their use is surprising given the usually occluded position of CAMs on the cell surface, such as at tight junctions. Nonetheless, the reason for their widespread involvement in virus entry most probably originates in their functional rather than structural characteristics.
Keywords: cell adhesion; cell entry; cryo-electron microscopy; receptor; virus.
Figures

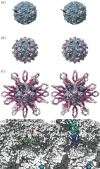
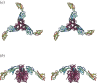
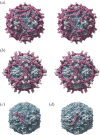
Similar articles
-
Virus-Receptor Interactions: The Key to Cellular Invasion.J Mol Biol. 2018 Aug 17;430(17):2590-2611. doi: 10.1016/j.jmb.2018.06.024. Epub 2018 Jun 18. J Mol Biol. 2018. PMID: 29924965 Free PMC article. Review.
-
Attachment factors.Adv Exp Med Biol. 2013;790:1-23. doi: 10.1007/978-1-4614-7651-1_1. Adv Exp Med Biol. 2013. PMID: 23884583 Review.
-
Different roles of the three loops forming the adhesive interface of nectin-4 in measles virus binding and cell entry, nectin-4 homodimerization, and heterodimerization with nectin-1.J Virol. 2014 Dec;88(24):14161-71. doi: 10.1128/JVI.02379-14. Epub 2014 Oct 1. J Virol. 2014. PMID: 25275122 Free PMC article.
-
CADM1 and CADM2 Trigger Neuropathogenic Measles Virus-Mediated Membrane Fusion by Acting in cis.J Virol. 2021 Jun 24;95(14):e0052821. doi: 10.1128/JVI.00528-21. Epub 2021 Jun 24. J Virol. 2021. PMID: 33910952 Free PMC article.
-
Conserved Surface Residues on the Feline Calicivirus Capsid Are Essential for Interaction with Its Receptor Feline Junctional Adhesion Molecule A (fJAM-A).J Virol. 2018 Mar 28;92(8):e00035-18. doi: 10.1128/JVI.00035-18. Print 2018 Apr 15. J Virol. 2018. PMID: 29386293 Free PMC article.
Cited by
-
Broad-Spectrum Extracellular Antiviral Properties of Cucurbit[n]urils.ACS Infect Dis. 2022 Oct 14;8(10):2084-2095. doi: 10.1021/acsinfecdis.2c00186. Epub 2022 Sep 5. ACS Infect Dis. 2022. PMID: 36062478 Free PMC article.
-
COVID-19, Renin-Angiotensin System and Endothelial Dysfunction.Cells. 2020 Jul 9;9(7):1652. doi: 10.3390/cells9071652. Cells. 2020. PMID: 32660065 Free PMC article. Review.
-
Whole Transcriptome Analysis of Aedes albopictus Mosquito Head and Thorax Post-Chikungunya Virus Infection.Pathogens. 2019 Aug 27;8(3):132. doi: 10.3390/pathogens8030132. Pathogens. 2019. PMID: 31461898 Free PMC article.
-
Polarized AAVR expression determines infectivity by AAV gene therapy vectors.Gene Ther. 2019 Jun;26(6):240-249. doi: 10.1038/s41434-019-0078-3. Epub 2019 Apr 8. Gene Ther. 2019. PMID: 30962536 Free PMC article.
-
Rhinoviruses and Their Receptors: Implications for Allergic Disease.Curr Allergy Asthma Rep. 2016 Apr;16(4):30. doi: 10.1007/s11882-016-0608-7. Curr Allergy Asthma Rep. 2016. PMID: 26960297 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

