Genotype to phenotype relationships in autism spectrum disorders
- PMID: 25531569
- PMCID: PMC4397214
- DOI: 10.1038/nn.3907
Genotype to phenotype relationships in autism spectrum disorders
Abstract
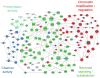
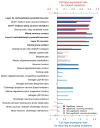
Autism spectrum disorders (ASDs) are characterized by phenotypic and genetic heterogeneity. Our analysis of functional networks perturbed in ASD suggests that both truncating and nontruncating de novo mutations contribute to autism, with a bias against truncating mutations in early embryonic development. We find that functional mutations are preferentially observed in genes likely to be haploinsufficient. Multiple cell types and brain areas are affected, but the impact of ASD mutations appears to be strongest in cortical interneurons, pyramidal neurons and the medium spiny neurons of the striatum, implicating cortical and corticostriatal brain circuits. In females, truncating ASD mutations on average affect genes with 50-100% higher brain expression than in males. Our results also suggest that truncating de novo mutations play a smaller role in the etiology of high-functioning ASD cases. Overall, we find that stronger functional insults usually lead to more severe intellectual, social and behavioral ASD phenotypes.
Figures

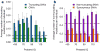
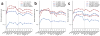
Similar articles
-
Exons as units of phenotypic impact for truncating mutations in autism.Mol Psychiatry. 2021 May;26(5):1685-1695. doi: 10.1038/s41380-020-00876-3. Epub 2020 Oct 27. Mol Psychiatry. 2021. PMID: 33110259
-
Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting.Brain Res. 2011 Mar 22;1380:42-77. doi: 10.1016/j.brainres.2010.11.078. Epub 2010 Dec 1. Brain Res. 2011. PMID: 21129364 Review.
-
Increased burden of deleterious variants in essential genes in autism spectrum disorder.Proc Natl Acad Sci U S A. 2016 Dec 27;113(52):15054-15059. doi: 10.1073/pnas.1613195113. Epub 2016 Dec 12. Proc Natl Acad Sci U S A. 2016. PMID: 27956632 Free PMC article.
-
Allelic expression analysis in the brain suggests a role for heterogeneous insults affecting epigenetic processes in autism spectrum disorders.Hum Mol Genet. 2014 Aug 1;23(15):4111-24. doi: 10.1093/hmg/ddu128. Epub 2014 Mar 21. Hum Mol Genet. 2014. PMID: 24659497
-
Recent developments in the genetics of autism spectrum disorders.Curr Opin Genet Dev. 2013 Jun;23(3):310-5. doi: 10.1016/j.gde.2013.02.003. Epub 2013 Mar 25. Curr Opin Genet Dev. 2013. PMID: 23537858 Review.
Cited by
-
Autistic traits foster effective curiosity-driven exploration.PLoS Comput Biol. 2024 Oct 31;20(10):e1012453. doi: 10.1371/journal.pcbi.1012453. eCollection 2024 Oct. PLoS Comput Biol. 2024. PMID: 39480751 Free PMC article.
-
A model-driven methodology for exploring complex disease comorbidities applied to autism spectrum disorder and inflammatory bowel disease.J Biomed Inform. 2016 Oct;63:366-378. doi: 10.1016/j.jbi.2016.08.008. Epub 2016 Aug 10. J Biomed Inform. 2016. PMID: 27522000 Free PMC article.
-
Unveiling the Pathogenesis of Psychiatric Disorders Using Network Models.Genes (Basel). 2021 Jul 20;12(7):1101. doi: 10.3390/genes12071101. Genes (Basel). 2021. PMID: 34356117 Free PMC article. Review.
-
Strength of functional signature correlates with effect size in autism.Genome Med. 2017 Jul 7;9(1):64. doi: 10.1186/s13073-017-0455-8. Genome Med. 2017. PMID: 28687074 Free PMC article.
-
Heterogeneity of autism symptoms in community-referred infants and toddlers at elevated or low familial likelihood of autism.Autism Res. 2023 Sep;16(9):1739-1749. doi: 10.1002/aur.2973. Epub 2023 Jul 5. Autism Res. 2023. PMID: 37408377 Free PMC article.
References
-
- Freitag CM. The genetics of autistic disorders and its clinical relevance: a review of the literature. Molecular psychiatry. 2007;12:2–22. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

