In vivo activation of a conserved microRNA program induces mammalian heart regeneration
- PMID: 25517466
- PMCID: PMC4270016
- DOI: 10.1016/j.stem.2014.10.003
In vivo activation of a conserved microRNA program induces mammalian heart regeneration
Erratum in
- Cell Stem Cell. 2014 Dec 4;15(6):805
Abstract
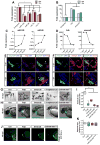
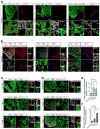
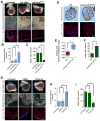
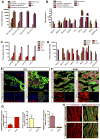
Heart failure is a leading cause of mortality and morbidity in the developed world, partly because mammals lack the ability to regenerate heart tissue. Whether this is due to evolutionary loss of regenerative mechanisms present in other organisms or to an inability to activate such mechanisms is currently unclear. Here we decipher mechanisms underlying heart regeneration in adult zebrafish and show that the molecular regulators of this response are conserved in mammals. We identified miR-99/100 and Let-7a/c and their protein targets smarca5 and fntb as critical regulators of cardiomyocyte dedifferentiation and heart regeneration in zebrafish. Although human and murine adult cardiomyocytes fail to elicit an endogenous regenerative response after myocardial infarction, we show that in vivo manipulation of this molecular machinery in mice results in cardiomyocyte dedifferentiation and improved heart functionality after injury. These data provide a proof of concept for identifying and activating conserved molecular programs to regenerate the damaged heart.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
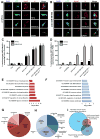
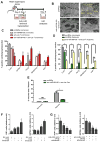
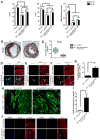
Comment in
-
Conserved microRNA program as key to mammalian cardiac regeneration: insights from zebrafish.Circ Res. 2015 Mar 27;116(7):1109-11. doi: 10.1161/CIRCRESAHA.115.305852. Circ Res. 2015. PMID: 25814680 No abstract available.
Similar articles
-
Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation.Nature. 2010 Mar 25;464(7288):606-9. doi: 10.1038/nature08899. Nature. 2010. PMID: 20336145 Free PMC article.
-
Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish.Elife. 2015 Apr 1;4:e05871. doi: 10.7554/eLife.05871. Elife. 2015. PMID: 25830562 Free PMC article.
-
Myocardial NF-κB activation is essential for zebrafish heart regeneration.Proc Natl Acad Sci U S A. 2015 Oct 27;112(43):13255-60. doi: 10.1073/pnas.1511209112. Epub 2015 Oct 15. Proc Natl Acad Sci U S A. 2015. PMID: 26472034 Free PMC article.
-
Cellular and Molecular Mechanism of Cardiac Regeneration: A Comparison of Newts, Zebrafish, and Mammals.Biomolecules. 2020 Aug 19;10(9):1204. doi: 10.3390/biom10091204. Biomolecules. 2020. PMID: 32825069 Free PMC article. Review.
-
Endogenous Mechanisms of Cardiac Regeneration.Int Rev Cell Mol Biol. 2016;326:67-131. doi: 10.1016/bs.ircmb.2016.04.002. Epub 2016 Jul 14. Int Rev Cell Mol Biol. 2016. PMID: 27572127 Review.
Cited by
-
Cardiac Regeneration After Myocardial Infarction: an Approachable Goal.Curr Cardiol Rep. 2020 Aug 10;22(10):122. doi: 10.1007/s11886-020-01361-7. Curr Cardiol Rep. 2020. PMID: 32778947 Free PMC article. Review.
-
Simulated Microgravity Exerts an Age-Dependent Effect on the Differentiation of Cardiovascular Progenitors Isolated from the Human Heart.PLoS One. 2015 Jul 10;10(7):e0132378. doi: 10.1371/journal.pone.0132378. eCollection 2015. PLoS One. 2015. PMID: 26161778 Free PMC article.
-
Mature miR-99a Upregulation in the Amniotic Fluid Samples from Female Fetus Down Syndrome Pregnancies: A Pilot Study.Medicina (Kaunas). 2019 Nov 7;55(11):728. doi: 10.3390/medicina55110728. Medicina (Kaunas). 2019. PMID: 31703316 Free PMC article.
-
Ablation of a Single N-Glycosylation Site in Human FSTL 1 Induces Cardiomyocyte Proliferation and Cardiac Regeneration.Mol Ther Nucleic Acids. 2018 Dec 7;13:133-143. doi: 10.1016/j.omtn.2018.08.021. Epub 2018 Sep 1. Mol Ther Nucleic Acids. 2018. PMID: 30290305 Free PMC article.
-
Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction.Nat Commun. 2019 Apr 17;10(1):1802. doi: 10.1038/s41467-019-09530-1. Nat Commun. 2019. PMID: 30996254 Free PMC article.
References
-
- Abad M, Mosteiro L, Pantoja C, Cañamero M, Rayon T, Ors I, Graña O, Megías D, Domínguez O, Martínez D, et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–345. - PubMed
-
- Aguirre A, Sancho-Martinez I, Izpisua Belmonte JC. Reprogramming toward heart regeneration: stem cells and beyond. Cell Stem Cell. 2013;12:275–284. - PubMed
-
- Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

