SIRT1 deacetylates TopBP1 and modulates intra-S-phase checkpoint and DNA replication origin firing
- PMID: 25516717
- PMCID: PMC4261203
- DOI: 10.7150/ijbs.11066
SIRT1 deacetylates TopBP1 and modulates intra-S-phase checkpoint and DNA replication origin firing
Abstract
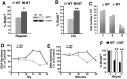
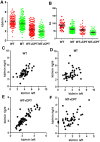
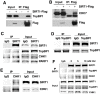
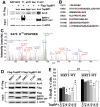
SIRT1, the mammalian homolog of yeast Sir2, is a founding member of a family of 7 protein and histone deacetylases that are involved in numerous biological functions. Previous studies revealed that SIRT1 deficiency results in genome instability, which eventually leads to cancer formation, yet the underlying mechanism is unclear. To investigate this, we conducted a proteomics study and found that SIRT1 interacted with many proteins involved in replication fork protection and origin firing. We demonstrated that loss of SIRT1 resulted in increased replication origin firing, asymmetric fork progression, defective intra-S-phase checkpoint, and chromosome damage. Mechanistically, SIRT1 deacetylates and affects the activity of TopBP1, which plays an essential role in DNA replication fork protection and replication origin firing. Our study demonstrated that ectopic over-expression of the deacetylated form of TopBP1 in SIRT1 mutant cells repressed replication origin firing, while the acetylated form of TopBP1 lost this function. Thus, SIRT1 acts upstream of TopBP1 and plays an essential role in maintaining genome stability by modulating DNA replication fork initiation and the intra-S-phase cell cycle checkpoint.
Keywords: DNA replication fork; Genetic stability.; Intra-S-phase checkpoint; SIRT1; TopBP1.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
A divergent role of the SIRT1-TopBP1 axis in regulating metabolic checkpoint and DNA damage checkpoint.Mol Cell. 2014 Dec 4;56(5):681-95. doi: 10.1016/j.molcel.2014.10.007. Epub 2014 Nov 13. Mol Cell. 2014. PMID: 25454945 Free PMC article.
-
Phosphorylated SIRT1 associates with replication origins to prevent excess replication initiation and preserve genomic stability.Nucleic Acids Res. 2017 Jul 27;45(13):7807-7824. doi: 10.1093/nar/gkx468. Nucleic Acids Res. 2017. PMID: 28549174 Free PMC article.
-
The ATR-Activation Domain of TopBP1 Is Required for the Suppression of Origin Firing during the S Phase.Int J Mol Sci. 2018 Aug 13;19(8):2376. doi: 10.3390/ijms19082376. Int J Mol Sci. 2018. PMID: 30104465 Free PMC article.
-
Extra View: Sirt1 Acts As A Gatekeeper Of Replication Initiation To Preserve Genomic Stability.Nucleus. 2018 Jan 1;9(1):261-267. doi: 10.1080/19491034.2018.1456218. Nucleus. 2018. PMID: 29578371 Free PMC article. Review.
-
The Intra-S Checkpoint Responses to DNA Damage.Genes (Basel). 2017 Feb 17;8(2):74. doi: 10.3390/genes8020074. Genes (Basel). 2017. PMID: 28218681 Free PMC article. Review.
Cited by
-
p300 arrests intervertebral disc degeneration by regulating the FOXO3/Sirt1/Wnt/β-catenin axis.Aging Cell. 2022 Aug;21(8):e13677. doi: 10.1111/acel.13677. Epub 2022 Jul 30. Aging Cell. 2022. PMID: 35907249 Free PMC article.
-
CK2 Phosphorylation of Human Papillomavirus 16 E2 on Serine 23 Promotes Interaction with TopBP1 and Is Critical for E2 Interaction with Mitotic Chromatin and the Viral Life Cycle.mBio. 2021 Oct 26;12(5):e0116321. doi: 10.1128/mBio.01163-21. Epub 2021 Sep 21. mBio. 2021. PMID: 34544280 Free PMC article.
-
Preventing excess replication origin activation to ensure genome stability.Trends Genet. 2022 Feb;38(2):169-181. doi: 10.1016/j.tig.2021.09.008. Epub 2021 Oct 6. Trends Genet. 2022. PMID: 34625299 Free PMC article. Review.
-
ATRIP Deacetylation by SIRT2 Drives ATR Checkpoint Activation by Promoting Binding to RPA-ssDNA.Cell Rep. 2016 Feb 16;14(6):1435-1447. doi: 10.1016/j.celrep.2016.01.018. Epub 2016 Feb 4. Cell Rep. 2016. PMID: 26854234 Free PMC article.
-
Convergence of SIRT1 and ATR signaling to modulate replication origin dormancy.Nucleic Acids Res. 2022 May 20;50(9):5111-5128. doi: 10.1093/nar/gkac299. Nucleic Acids Res. 2022. PMID: 35524559 Free PMC article.
References
-
- Boye E, Skjolberg HC, Grallert B. Checkpoint regulation of DNA replication. Methods in molecular biology. 2009;521:55–70. - PubMed
-
- Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Critical reviews in biochemistry and molecular biology. 2012;47:222–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

