Defective hydrophobic sliding mechanism and active site expansion in HIV-1 protease drug resistant variant Gly48Thr/Leu89Met: mechanisms for the loss of saquinavir binding potency
- PMID: 25513833
- PMCID: PMC4303317
- DOI: 10.1021/bi501088e
Defective hydrophobic sliding mechanism and active site expansion in HIV-1 protease drug resistant variant Gly48Thr/Leu89Met: mechanisms for the loss of saquinavir binding potency
Abstract
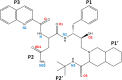
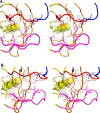
HIV drug resistance continues to emerge; consequently, there is an urgent need to develop next generation antiretroviral therapeutics.1 Here we report on the structural and kinetic effects of an HIV protease drug resistant variant with the double mutations Gly48Thr and Leu89Met (PRG48T/L89M), without the stabilizing mutations Gln7Lys, Leu33Ile, and Leu63Ile. Kinetic analyses reveal that PRG48T/L89M and PRWT share nearly identical Michaelis-Menten parameters; however, PRG48T/L89M exhibits weaker binding for IDV (41-fold), SQV (18-fold), APV (15-fold), and NFV (9-fold) relative to PRWT. A 1.9 Å resolution crystal structure was solved for PRG48T/L89M bound with saquinavir (PRG48T/L89M-SQV) and compared to the crystal structure of PRWT bound with saquinavir (PRWT-SQV). PRG48T/L89M-SQV has an enlarged active site resulting in the loss of a hydrogen bond in the S3 subsite from Gly48 to P3 of SQV, as well as less favorable hydrophobic packing interactions between P1 Phe of SQV and the S1 subsite. PRG48T/L89M-SQV assumes a more open conformation relative to PRWT-SQV, as illustrated by the downward displacement of the fulcrum and elbows and weaker interatomic flap interactions. We also show that the Leu89Met mutation disrupts the hydrophobic sliding mechanism by causing a redistribution of van der Waals interactions in the hydrophobic core in PRG48T/L89M-SQV. Our mechanism for PRG48T/L89M-SQV drug resistance proposes that a defective hydrophobic sliding mechanism results in modified conformational dynamics of the protease. As a consequence, the protease is unable to achieve a fully closed conformation that results in an expanded active site and weaker inhibitor binding.
Figures
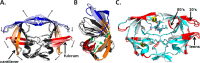



Similar articles
-
Structural and kinetic analyses of the protease from an amprenavir-resistant human immunodeficiency virus type 1 mutant rendered resistant to saquinavir and resensitized to amprenavir.J Virol. 2000 Aug;74(16):7636-41. doi: 10.1128/jvi.74.16.7636-7641.2000. J Virol. 2000. PMID: 10906218 Free PMC article.
-
Structural Basis of Why Nelfinavir-Resistant D30N Mutant of HIV-1 Protease Remains Susceptible to Saquinavir.Chem Biol Drug Des. 2015 Sep;86(3):302-8. doi: 10.1111/cbdd.12494. Epub 2015 Jan 9. Chem Biol Drug Des. 2015. PMID: 25487655
-
Structural analysis of an HIV-1 protease I47A mutant resistant to the protease inhibitor lopinavir.Protein Sci. 2005 Jul;14(7):1870-8. doi: 10.1110/ps.051347405. Epub 2005 Jun 3. Protein Sci. 2005. PMID: 15937277 Free PMC article.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
-
Structural and thermodynamic basis of resistance to HIV-1 protease inhibition: implications for inhibitor design.Curr Drug Targets Infect Disord. 2003 Dec;3(4):311-28. doi: 10.2174/1568005033481051. Curr Drug Targets Infect Disord. 2003. PMID: 14754432 Review.
Cited by
-
Characterizing early drug resistance-related events using geometric ensembles from HIV protease dynamics.Sci Rep. 2018 Dec 18;8(1):17938. doi: 10.1038/s41598-018-36041-8. Sci Rep. 2018. PMID: 30560871 Free PMC article.
-
Improving Viral Protease Inhibitors to Counter Drug Resistance.Trends Microbiol. 2016 Jul;24(7):547-557. doi: 10.1016/j.tim.2016.03.010. Epub 2016 Apr 15. Trends Microbiol. 2016. PMID: 27090931 Free PMC article. Review.
-
Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.FEBS J. 2020 Aug;287(15):3235-3254. doi: 10.1111/febs.15207. Epub 2020 Jan 23. FEBS J. 2020. PMID: 31920003 Free PMC article.
-
Natural Polymorphisms D60E and I62V Stabilize a Closed Conformation in HIV-1 Protease in the Absence of an Inhibitor or Substrate.Viruses. 2024 Feb 2;16(2):236. doi: 10.3390/v16020236. Viruses. 2024. PMID: 38400012 Free PMC article.
-
Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.J Biol Chem. 2016 Oct 21;291(43):22741-22756. doi: 10.1074/jbc.M116.747568. Epub 2016 Aug 30. J Biol Chem. 2016. PMID: 27576689 Free PMC article.
References
-
- World Health Organization, UNAIDS (March 2014) Surveilllance of HIV Drug Resistance in Adults Initiating Antiretroviral Therapy (Pretreatment HIV Drug Resistance), http://www.who.int/hiv/pub/drugresistance/pretreatment_drugresistance/en/.
-
- World Health Organization (2013) Number of deaths due to HIV/AIDS; http://www.who.int/gho/hiv/epidemic_status/deaths_text/en/.
-
- Tozser J. (2001) HIV Inhibitors: Problems and Reality. Ann. N.Y. Acad. Sci. 946, 145–159. - PubMed
-
- Hammer S. M.; Squires K. E.; Hughes M. D.; Grimes J. M.; Demeter L. M.; Currier J. S.; Eron J. J. Jr.; Feinberg J. E.; Balfour H. H. Jr.; Deyton L. R.; Chodakewitz J. A.; Fischl M. A. (1997) A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 337, 725–733. - PubMed
-
- Condra J. H.; Schleif W. A.; Blahy O. M.; Gabryelski L. J.; Graham D. J.; Quintero J. C.; Rhodes A.; Robbins H. L.; Roth E.; Shivaprakash M. (1995) In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374, 569–571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

