From cells to muropeptide structures in 24 h: peptidoglycan mapping by UPLC-MS
- PMID: 25510564
- PMCID: PMC4267204
- DOI: 10.1038/srep07494
From cells to muropeptide structures in 24 h: peptidoglycan mapping by UPLC-MS
Abstract
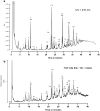
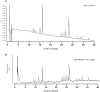
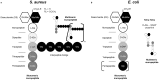
Peptidoglycan (PGN) is ubiquitous in nearly all bacterial species. The PGN sacculus protects the cells against their own internal turgor making PGN one of the most important targets for antibacterial treatment. Within the last sixty years PGN composition has been intensively studied by various methods. The breakthrough was the application of HPLC technology on the analysis of muropeptides. However, preparation of pure PGN relied on a very time consuming method of about one week. We established a purification protocol for both Gram-positive and Gram-negative bacteria which can be completely performed in plastic reaction tubes yielding pure muropeptides within 24 hours. The muropeptides can be analyzed by UPLC-MS, allowing their immediate determination. This new rapid method provides the feasibility to screen PGN composition even in high throughput, making it a highly useful tool for basic research as well as for the pharmaceutical industry.
Figures
Similar articles
-
Negative and positive ion matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and positive ion nano-electrospray ionization quadrupole ion trap mass spectrometry of peptidoglycan fragments isolated from various Bacillus species.J Mass Spectrom. 2001 Feb;36(2):124-39. doi: 10.1002/jms.109. J Mass Spectrom. 2001. PMID: 11288194
-
Ultra-Sensitive, High-Resolution Liquid Chromatography Methods for the High-Throughput Quantitative Analysis of Bacterial Cell Wall Chemistry and Structure.Methods Mol Biol. 2016;1440:11-27. doi: 10.1007/978-1-4939-3676-2_2. Methods Mol Biol. 2016. PMID: 27311661
-
Use of Capillary Zone Electrophoresis Coupled to Electrospray Mass Spectrometry for the Detection and Absolute Quantitation of Peptidoglycan-Derived Peptides in Bacterial Cytoplasmic Extracts.Anal Chem. 2021 Feb 2;93(4):2342-2350. doi: 10.1021/acs.analchem.0c04218. Epub 2021 Jan 20. Anal Chem. 2021. PMID: 33470796
-
Peptidoglycan perception--sensing bacteria by their common envelope structure.Int J Med Microbiol. 2015 Feb;305(2):217-23. doi: 10.1016/j.ijmm.2014.12.019. Epub 2014 Dec 24. Int J Med Microbiol. 2015. PMID: 25596887 Review.
-
Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection.J Innate Immun. 2009;1(2):88-97. doi: 10.1159/000181181. J Innate Immun. 2009. PMID: 19319201 Free PMC article. Review.
Cited by
-
Class A Penicillin-Binding Protein C Is Responsible for Stress Response by Regulation of Peptidoglycan Assembly in Clavibacter michiganensis.Microbiol Spectr. 2022 Oct 26;10(5):e0181622. doi: 10.1128/spectrum.01816-22. Epub 2022 Aug 30. Microbiol Spectr. 2022. PMID: 36040162 Free PMC article.
-
An updated toolkit for exploring bacterial cell wall structure and dynamics.Fac Rev. 2021 Feb 10;10:14. doi: 10.12703/r/10-14. eCollection 2021. Fac Rev. 2021. PMID: 33659932 Free PMC article. Review.
-
The serine/threonine kinase Stk and the phosphatase Stp regulate cell wall synthesis in Staphylococcus aureus.Sci Rep. 2018 Sep 12;8(1):13693. doi: 10.1038/s41598-018-32109-7. Sci Rep. 2018. PMID: 30209409 Free PMC article.
-
Staphylococcus aureus susceptibility to complestatin and corbomycin depends on the VraSR two-component system.Microbiol Spectr. 2023 Aug 30;11(5):e0037023. doi: 10.1128/spectrum.00370-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37646518 Free PMC article.
-
Tumescenamide C, a cyclic lipodepsipeptide from Streptomyces sp. KUSC_F05, exerts antimicrobial activity against the scab-forming actinomycete Streptomyces scabiei.J Antibiot (Tokyo). 2024 Jun;77(6):353-364. doi: 10.1038/s41429-024-00716-4. Epub 2024 Mar 25. J Antibiot (Tokyo). 2024. PMID: 38523145
References
-
- Salton M. R. J. & Horne R. W. Studies of the bacterial cell wall. 2. Methods of preparation and some properties of cell walls. Biochimica et Biophysica Acta 7, 177–197 (1951). - PubMed
-
- Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172, 451–464 (1988). - PubMed
-
- de Jonge B. L., Chang Y. S., Gage D. & Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem 267, 11248–11254 (1992). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

