Nitrogen regulation of fungal secondary metabolism in fungi
- PMID: 25506342
- PMCID: PMC4246892
- DOI: 10.3389/fmicb.2014.00656
Nitrogen regulation of fungal secondary metabolism in fungi
Abstract
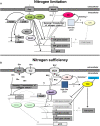
Fungi occupy diverse environments where they are constantly challenged by stressors such as extreme pH, temperature, UV exposure, and nutrient deprivation. Nitrogen is an essential requirement for growth, and the ability to metabolize a wide variety of nitrogen sources enables fungi to colonize different environmental niches and survive nutrient limitations. Favored nitrogen sources, particularly ammonium and glutamine, are used preferentially, while the expression of genes required for the use of various secondary nitrogen sources is subject to a regulatory mechanism called nitrogen metabolite repression. Studies on gene regulation in response to nitrogen availability were carried out first in Saccharomyces cerevisiae, Aspergillus nidulans, and Neurospora crassa. These studies revealed that fungi respond to changes in nitrogen availability with physiological and morphological alterations and activation of differentiation processes. In all fungal species studied, the major GATA transcription factor AreA and its co-repressor Nmr are central players of the nitrogen regulatory network. In addition to growth and development, the quality and quantity of nitrogen also affects the formation of a broad range of secondary metabolites (SMs). Recent studies, mainly on species of the genus Fusarium, revealed that AreA does not only regulate a large set of nitrogen catabolic genes, but can also be involved in regulating production of SMs. Furthermore, several other regulators, e.g., a second GATA transcription factor, AreB, that was proposed to negatively control nitrogen catabolic genes by competing with AreA for binding to GATA elements, was shown to act as activator of some nitrogen-repressed as well as nitrogen-induced SM gene clusters. This review highlights our latest understanding of canonical (AreA-dependent) and non-canonical nitrogen regulation mechanisms by which fungi may regulate biosynthesis of certain SMs in response to nitrogen availability.
Keywords: AreA; AreB; GS; MeaB; Nmr1; nitrogen regulation; secondary metabolites.
Figures
Similar articles
-
The interplay between the GATA transcription factors AreA, the global nitrogen regulator and AreB in Fusarium fujikuroi.Mol Microbiol. 2014 Feb;91(3):472-93. doi: 10.1111/mmi.12472. Epub 2013 Dec 19. Mol Microbiol. 2014. PMID: 24286256
-
Comparative transcriptome and proteome analysis reveals a global impact of the nitrogen regulators AreA and AreB on secondary metabolism in Fusarium fujikuroi.PLoS One. 2017 Apr 25;12(4):e0176194. doi: 10.1371/journal.pone.0176194. eCollection 2017. PLoS One. 2017. PMID: 28441411 Free PMC article.
-
AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR.Mol Microbiol. 2003 Feb;47(4):975-91. doi: 10.1046/j.1365-2958.2003.03326.x. Mol Microbiol. 2003. PMID: 12581353
-
Genetic regulation of nitrogen metabolism in the fungi.Microbiol Mol Biol Rev. 1997 Mar;61(1):17-32. doi: 10.1128/mmbr.61.1.17-32.1997. Microbiol Mol Biol Rev. 1997. PMID: 9106362 Free PMC article. Review.
-
Nitrogen regulation of virulence in clinically prevalent fungal pathogens.FEMS Microbiol Lett. 2013 Aug;345(2):77-84. doi: 10.1111/1574-6968.12181. Epub 2013 Jun 13. FEMS Microbiol Lett. 2013. PMID: 23701678 Review.
Cited by
-
Current progress on pathogenicity-related transcription factors in Fusarium oxysporum.Mol Plant Pathol. 2021 Jul;22(7):882-895. doi: 10.1111/mpp.13068. Epub 2021 May 9. Mol Plant Pathol. 2021. PMID: 33969616 Free PMC article. Review.
-
Aspergillus nidulans: A Potential Resource of the Production of the Native and Heterologous Enzymes for Industrial Applications.Int J Microbiol. 2020 Aug 1;2020:8894215. doi: 10.1155/2020/8894215. eCollection 2020. Int J Microbiol. 2020. PMID: 32802076 Free PMC article. Review.
-
Penicillium chrysogenum, a Vintage Model with a Cutting-Edge Profile in Biotechnology.Microorganisms. 2022 Mar 6;10(3):573. doi: 10.3390/microorganisms10030573. Microorganisms. 2022. PMID: 35336148 Free PMC article. Review.
-
Secondary metabolites in fungus-plant interactions.Front Plant Sci. 2015 Aug 6;6:573. doi: 10.3389/fpls.2015.00573. eCollection 2015. Front Plant Sci. 2015. PMID: 26300892 Free PMC article. Review.
-
GCN4 Enhances the Transcriptional Regulation of AreA by Interacting with SKO1 To Mediate Nitrogen Utilization in Ganoderma lucidum.Appl Environ Microbiol. 2022 Nov 22;88(22):e0132222. doi: 10.1128/aem.01322-22. Epub 2022 Nov 7. Appl Environ Microbiol. 2022. PMID: 36342130 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

