TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA
- PMID: 25503365
- PMCID: PMC4284588
- DOI: 10.1073/pnas.1413994112
TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA
Abstract
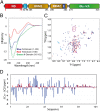
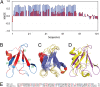
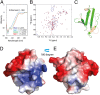
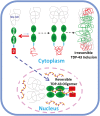
Transactivation response element (TAR) DNA-binding protein 43 (TDP-43) is the principal component of ubiquitinated inclusions characteristic of most forms of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia-frontotemporal lobar degeneration with TDP-43-positive inclusions (FTLD-TDP), as well as an increasing spectrum of other neurodegenerative diseases. Previous structural and functional studies on TDP-43 have been mostly focused on its recognized domains. Very recently, however, its extreme N terminus was identified to be a double-edged sword indispensable for both physiology and proteinopathy, but thus far its structure remains unknown due to the severe aggregation. Here as facilitated by our previous discovery that protein aggregation can be significantly minimized by reducing salt concentrations, by circular dichroism and NMR spectroscopy we revealed that the TDP-43 N terminus encodes a well-folded structure in concentration-dependent equilibrium with its unfolded form. Despite previous failure in detecting any sequence homology to ubiquitin, the folded state was determined to adopt a novel ubiquitin-like fold by the CS-Rosetta program with NMR chemical shifts and 78 unambiguous long-range nuclear Overhauser effect (NOE) constraints. Remarkably, this ubiquitin-like fold could bind ssDNA, and the binding shifted the conformational equilibrium toward reducing the unfolded population. To the best of our knowledge, the TDP-43 N terminus represents the first ubiquitin-like fold capable of directly binding nucleic acid. Our results provide a molecular mechanism rationalizing the functional dichotomy of TDP-43 and might also shed light on the formation and dynamics of cellular ribonucleoprotein granules, which have been recently linked to ALS pathogenesis. As a consequence, one therapeutic strategy for TDP-43-causing diseases might be to stabilize its ubiquitin-like fold by ssDNA or designed molecules.
Keywords: FTLD-TDP; NMR spectroscopy; TDP-43; amyotrophic lateral sclerosis; ubiquitin-like fold.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity.Biochem Biophys Res Commun. 2012 Aug 24;425(2):219-24. doi: 10.1016/j.bbrc.2012.07.071. Epub 2012 Jul 23. Biochem Biophys Res Commun. 2012. PMID: 22835933
-
Inter-domain interactions of TDP-43 as decoded by NMR.Biochem Biophys Res Commun. 2016 Apr 29;473(2):614-9. doi: 10.1016/j.bbrc.2016.03.158. Epub 2016 Apr 1. Biochem Biophys Res Commun. 2016. PMID: 27040765
-
Folding of the RNA recognition motif (RRM) domains of the amyotrophic lateral sclerosis (ALS)-linked protein TDP-43 reveals an intermediate state.J Biol Chem. 2014 Mar 21;289(12):8264-76. doi: 10.1074/jbc.M113.542779. Epub 2014 Feb 4. J Biol Chem. 2014. PMID: 24497641 Free PMC article.
-
[Component of ubiquitin-positive inclusions in ALS].Brain Nerve. 2007 Oct;59(10):1171-7. Brain Nerve. 2007. PMID: 17969358 Review. Japanese.
-
The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia.Curr Opin Neurol. 2008 Dec;21(6):693-700. doi: 10.1097/WCO.0b013e3283168d1d. Curr Opin Neurol. 2008. PMID: 18989115 Free PMC article. Review.
Cited by
-
Amyotrophic Lateral Sclerosis: Proteins, Proteostasis, Prions, and Promises.Front Cell Neurosci. 2020 Nov 4;14:581907. doi: 10.3389/fncel.2020.581907. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33328890 Free PMC article.
-
Computational Insights of Unfolding of N-Terminal Domain of TDP-43 Reveal the Conformational Heterogeneity in the Unfolding Pathway.Front Mol Neurosci. 2022 Apr 25;15:822863. doi: 10.3389/fnmol.2022.822863. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35548668 Free PMC article.
-
Molecular Mechanisms of TDP-43 Misfolding and Pathology in Amyotrophic Lateral Sclerosis.Front Mol Neurosci. 2019 Feb 14;12:25. doi: 10.3389/fnmol.2019.00025. eCollection 2019. Front Mol Neurosci. 2019. PMID: 30837838 Free PMC article. Review.
-
Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions.J Biol Chem. 2017 Jul 14;292(28):11992-12006. doi: 10.1074/jbc.M117.775965. Epub 2017 May 31. J Biol Chem. 2017. PMID: 28566288 Free PMC article.
-
Somatic TARDBP variants as a cause of semantic dementia.Brain. 2020 Dec 1;143(12):3827-3841. doi: 10.1093/brain/awaa317. Brain. 2020. PMID: 33155043 Free PMC article.
References
-
- Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351(3):602–611. - PubMed
-
- Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

