Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair
- PMID: 25491644
- PMCID: PMC4317648
- DOI: 10.1534/genetics.114.172361
Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair
Abstract
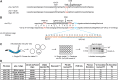
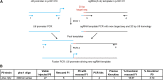
As in other organisms, CRISPR/Cas9 methods provide a powerful approach for genome editing in the nematode Caenorhabditis elegans. Oligonucleotides are excellent repair templates for introducing substitutions and short insertions, as they are cost effective, require no cloning, and appear in other organisms to target changes by homologous recombination at DNA double-strand breaks (DSBs). Here, I describe a methodology in C. elegans to efficiently knock in epitope tags in 8-9 days, using a temperature-sensitive lethal mutation in the pha-1 gene as a co-conversion marker. I demonstrate that 60mer oligos with 29 bp of homology drive efficient knock-in of point mutations, and that disabling nonhomologous end joining by RNAi inactivation of the cku-80 gene significantly improves knock-in efficiency. Homology arms of 35-80 bp are sufficient for efficient editing and DSBs up to 54 bp away from the insertion site produced knock-ins. These findings will likely be applicable for a range of genome editing approaches in C. elegans, which will improve editing efficiency and minimize screening efforts.
Keywords: CRISPR/Cas9; co-conversion; nonhomologous end joining; oligonucleotide-mediated homologous recombination; pha-1.
Copyright © 2015 by the Genetics Society of America.
Figures


Similar articles
-
The application of CRISPR-Cas9 genome editing in Caenorhabditis elegans.J Genet Genomics. 2015 Aug 20;42(8):413-21. doi: 10.1016/j.jgg.2015.06.005. Epub 2015 Jun 26. J Genet Genomics. 2015. PMID: 26336798 Free PMC article. Review.
-
CRISPR/Cas9 Genome Editing in Caenorhabditis elegans: Evaluation of Templates for Homology-Mediated Repair and Knock-Ins by Homology-Independent DNA Repair.G3 (Bethesda). 2015 Jun 3;5(8):1649-56. doi: 10.1534/g3.115.019273. G3 (Bethesda). 2015. PMID: 26044730 Free PMC article.
-
Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination.Nucleic Acids Res. 2013 Nov;41(20):e193. doi: 10.1093/nar/gkt805. Epub 2013 Sep 5. Nucleic Acids Res. 2013. PMID: 24013562 Free PMC article.
-
Highly efficient CRISPR/HDR-mediated knock-in for mouse embryonic stem cells and zygotes.Biotechniques. 2015 Oct 1;59(4):201-2, 204, 206-8. doi: 10.2144/000114339. eCollection 2015 Oct. Biotechniques. 2015. PMID: 26458548
-
Control of gene editing by manipulation of DNA repair mechanisms.Mamm Genome. 2017 Aug;28(7-8):262-274. doi: 10.1007/s00335-017-9688-5. Epub 2017 Apr 3. Mamm Genome. 2017. PMID: 28374058 Review.
Cited by
-
The conserved molting/circadian rhythm regulator NHR-23/NR1F1 serves as an essential co-regulator of C. elegans spermatogenesis.Development. 2020 Nov 27;147(22):dev193862. doi: 10.1242/dev.193862. Development. 2020. PMID: 33060131 Free PMC article.
-
Melting dsDNA Donor Molecules Greatly Improves Precision Genome Editing in Caenorhabditis elegans.Genetics. 2020 Nov;216(3):643-650. doi: 10.1534/genetics.120.303564. Epub 2020 Sep 22. Genetics. 2020. PMID: 32963112 Free PMC article.
-
Membrane-associated cytoplasmic granules carrying the Argonaute protein WAGO-3 enable paternal epigenetic inheritance in Caenorhabditis elegans.Nat Cell Biol. 2022 Feb;24(2):217-229. doi: 10.1038/s41556-021-00827-2. Epub 2022 Feb 7. Nat Cell Biol. 2022. PMID: 35132225 Free PMC article.
-
RAB-35 and ARF-6 GTPases Mediate Engulfment and Clearance Following Linker Cell-Type Death.Dev Cell. 2018 Oct 22;47(2):222-238.e6. doi: 10.1016/j.devcel.2018.08.015. Epub 2018 Sep 13. Dev Cell. 2018. PMID: 30220571 Free PMC article.
-
Mos1 Element-Mediated CRISPR Integration of Transgenes in Caenorhabditis elegans.G3 (Bethesda). 2019 Aug 8;9(8):2629-2635. doi: 10.1534/g3.119.400399. G3 (Bethesda). 2019. PMID: 31186306 Free PMC article.
References
-
- Adamo A., Collis S. J., Adelman C. A., Silva N., Horejsi Z., et al. , 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell 39: 25–35. - PubMed
-
- Asahina M., Ishihara T., Jindra M., Kohara Y., Katsura I., et al. , 2000. The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells 5: 711–723. - PubMed
-
- Asahina M., Valenta T., Silhánková M., Korinek V., Jindra M., 2006. Crosstalk between a nuclear receptor and beta-catenin signaling decides cell fates in the C. elegans somatic gonad. Dev. Cell 11: 203–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

