Automated nucleic acid chain tracing in real time
- PMID: 25485119
- PMCID: PMC4224457
- DOI: 10.1107/S2052252514019290
Automated nucleic acid chain tracing in real time
Abstract
The crystallographic structure solution of nucleotides and nucleotide complexes is now commonplace. The resulting electron-density maps are often poorer than for proteins, and as a result interpretation in terms of an atomic model can require significant effort, particularly in the case of large structures. While model building can be performed automatically, as with proteins, the process is time-consuming, taking minutes to days depending on the software and the size of the structure. A method is presented for the automatic building of nucleotide chains into electron density which is fast enough to be used in interactive model-building software, with extended chain fragments built around the current view position in a fraction of a second. The speed of the method arises from the determination of the 'fingerprint' of the sugar and phosphate groups in terms of conserved high-density and low-density features, coupled with a highly efficient scoring algorithm. Use cases include the rapid evaluation of an initial electron-density map, addition of nucleotide fragments to prebuilt protein structures, and in favourable cases the completion of the structure while automated model-building software is still running. The method has been incorporated into the Coot software package.
Keywords: Coot; nucleic acid chain tracing.
Figures
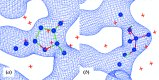
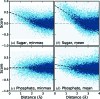
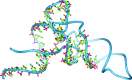
Similar articles
-
Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data.Protein Sci. 2020 Apr;29(4):1069-1078. doi: 10.1002/pro.3791. Epub 2020 Mar 2. Protein Sci. 2020. PMID: 31730249 Free PMC article.
-
Predicting protein model correctness in Coot using machine learning.Acta Crystallogr D Struct Biol. 2020 Aug 1;76(Pt 8):713-723. doi: 10.1107/S2059798320009080. Epub 2020 Jul 27. Acta Crystallogr D Struct Biol. 2020. PMID: 32744253 Free PMC article.
-
Use of noncrystallographic symmetry for automated model building at medium to low resolution.Acta Crystallogr D Biol Crystallogr. 2012 Apr;68(Pt 4):446-53. doi: 10.1107/S0907444911050712. Epub 2012 Mar 16. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22505265 Free PMC article.
-
De novo main-chain modeling for EM maps using MAINMAST.Nat Commun. 2018 Apr 24;9(1):1618. doi: 10.1038/s41467-018-04053-7. Nat Commun. 2018. PMID: 29691408 Free PMC article.
-
Interactive model building in neutron macromolecular crystallography.Methods Enzymol. 2020;634:201-224. doi: 10.1016/bs.mie.2019.11.017. Epub 2019 Dec 20. Methods Enzymol. 2020. PMID: 32093833 Review.
Cited by
-
De novo computational RNA modeling into cryo-EM maps of large ribonucleoprotein complexes.Nat Methods. 2018 Nov;15(11):947-954. doi: 10.1038/s41592-018-0172-2. Epub 2018 Oct 30. Nat Methods. 2018. PMID: 30377372 Free PMC article.
-
Strategies for carbohydrate model building, refinement and validation.Acta Crystallogr D Struct Biol. 2017 Feb 1;73(Pt 2):171-186. doi: 10.1107/S2059798316016910. Epub 2017 Feb 1. Acta Crystallogr D Struct Biol. 2017. PMID: 28177313 Free PMC article.
-
The Automatic Solution of Macromolecular Crystal Structures via Molecular Replacement Techniques: REMO22 and Its Pipeline.Int J Mol Sci. 2023 Mar 23;24(7):6070. doi: 10.3390/ijms24076070. Int J Mol Sci. 2023. PMID: 37047043 Free PMC article.
-
Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data.Protein Sci. 2020 Apr;29(4):1069-1078. doi: 10.1002/pro.3791. Epub 2020 Mar 2. Protein Sci. 2020. PMID: 31730249 Free PMC article.
-
EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.Nat Methods. 2015 Oct;12(10):943-6. doi: 10.1038/nmeth.3541. Epub 2015 Aug 17. Nat Methods. 2015. PMID: 26280328 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

