Cellular mechanisms of alpha herpesvirus egress: live cell fluorescence microscopy of pseudorabies virus exocytosis
- PMID: 25474634
- PMCID: PMC4256261
- DOI: 10.1371/journal.ppat.1004535
Cellular mechanisms of alpha herpesvirus egress: live cell fluorescence microscopy of pseudorabies virus exocytosis
Abstract
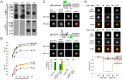
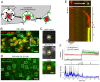
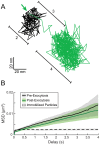
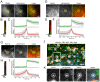
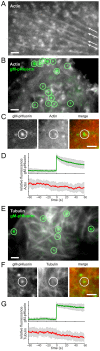
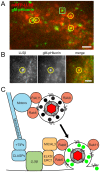
Egress of newly assembled herpesvirus particles from infected cells is a highly dynamic process involving the host secretory pathway working in concert with viral components. To elucidate the location, dynamics, and molecular mechanisms of alpha herpesvirus egress, we developed a live-cell fluorescence microscopy method to visualize the final transport and exocytosis of pseudorabies virus (PRV) particles in non-polarized epithelial cells. This method is based on total internal reflection fluorescence (TIRF) microscopy to selectively image fluorescent virus particles near the plasma membrane, and takes advantage of a virus-encoded pH-sensitive probe to visualize the precise moment and location of particle exocytosis. We performed single-particle tracking and mean squared displacement analysis to characterize particle motion, and imaged a panel of cellular proteins to identify those spatially and dynamically associated with viral exocytosis. Based on our data, individual virus particles travel to the plasma membrane inside small, acidified secretory vesicles. Rab GTPases, Rab6a, Rab8a, and Rab11a, key regulators of the plasma membrane-directed secretory pathway, are present on the virus secretory vesicle. These vesicles undergo fast, directional transport directly to the site of exocytosis, which is most frequently near patches of LL5β, part of a complex that anchors microtubules to the plasma membrane. Vesicles are tightly docked at the site of exocytosis for several seconds, and membrane fusion occurs, displacing the virion a small distance across the plasma membrane. After exocytosis, particles remain tightly confined on the outer cell surface. Based on recent reports in the cell biological and alpha herpesvirus literature, combined with our spatial and dynamic data on viral egress, we propose an integrated model that links together the intracellular transport pathways and exocytosis mechanisms that mediate alpha herpesvirus egress.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Herpes Simplex Virus 1 (HSV-1) Uses the Rab6 Post-Golgi Secretory Pathway For Viral Egress.bioRxiv [Preprint]. 2023 Dec 13:2023.12.13.571414. doi: 10.1101/2023.12.13.571414. bioRxiv. 2023. Update in: J Virol. 2024 Sep 17;98(9):e0059924. doi: 10.1128/jvi.00599-24. PMID: 38168379 Free PMC article. Updated. Preprint.
-
The Rab6 post-Golgi secretory pathway contributes to herpes simplex virus 1 (HSV-1) egress.J Virol. 2024 Sep 17;98(9):e0059924. doi: 10.1128/jvi.00599-24. Epub 2024 Aug 13. J Virol. 2024. PMID: 39136459 Free PMC article.
-
Exocytosis of Alphaherpesvirus Virions, Light Particles, and Glycoproteins Uses Constitutive Secretory Mechanisms.mBio. 2016 Jun 7;7(3):e00820-16. doi: 10.1128/mBio.00820-16. mBio. 2016. PMID: 27273828 Free PMC article.
-
The way out: what we know and do not know about herpesvirus nuclear egress.Cell Microbiol. 2013 Feb;15(2):170-8. doi: 10.1111/cmi.12044. Epub 2012 Nov 7. Cell Microbiol. 2013. PMID: 23057731 Review.
-
Role of Rab GTPases in HSV-1 infection: Molecular understanding of viral maturation and egress.Microb Pathog. 2018 May;118:146-153. doi: 10.1016/j.micpath.2018.03.028. Epub 2018 Mar 16. Microb Pathog. 2018. PMID: 29551438 Review.
Cited by
-
Herpes Simplex Virus 1 (HSV-1) Uses the Rab6 Post-Golgi Secretory Pathway For Viral Egress.bioRxiv [Preprint]. 2023 Dec 13:2023.12.13.571414. doi: 10.1101/2023.12.13.571414. bioRxiv. 2023. Update in: J Virol. 2024 Sep 17;98(9):e0059924. doi: 10.1128/jvi.00599-24. PMID: 38168379 Free PMC article. Updated. Preprint.
-
LIVE-CELL FLUORESCENCE MICROSCOPY OF HSV-1 CELLULAR EGRESS BY EXOCYTOSIS.bioRxiv [Preprint]. 2023 Aug 17:2023.02.27.530373. doi: 10.1101/2023.02.27.530373. bioRxiv. 2023. PMID: 36909512 Free PMC article. Preprint.
-
Alpha herpesvirus exocytosis from neuron cell bodies uses constitutive secretory mechanisms, and egress and spread from axons is independent of neuronal firing activity.PLoS Pathog. 2024 Apr 5;20(4):e1012139. doi: 10.1371/journal.ppat.1012139. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38578790 Free PMC article.
-
Experimental Dissection of the Lytic Replication Cycles of Herpes Simplex Viruses in vitro.Front Microbiol. 2018 Oct 11;9:2406. doi: 10.3389/fmicb.2018.02406. eCollection 2018. Front Microbiol. 2018. PMID: 30386309 Free PMC article. Review.
-
Transneuronal Circuit Analysis with Pseudorabies Viruses.Curr Protoc. 2023 Jul;3(7):e841. doi: 10.1002/cpz1.841. Curr Protoc. 2023. PMID: 37486157 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

