Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution
- PMID: 25470049
- PMCID: PMC4864027
- DOI: 10.1038/nature13952
Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution
Abstract
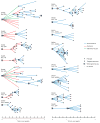
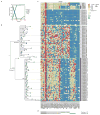
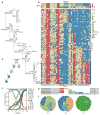
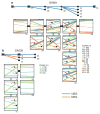
Human cancers, including breast cancers, comprise clones differing in mutation content. Clones evolve dynamically in space and time following principles of Darwinian evolution, underpinning important emergent features such as drug resistance and metastasis. Human breast cancer xenoengraftment is used as a means of capturing and studying tumour biology, and breast tumour xenografts are generally assumed to be reasonable models of the originating tumours. However, the consequences and reproducibility of engraftment and propagation on the genomic clonal architecture of tumours have not been systematically examined at single-cell resolution. Here we show, using deep-genome and single-cell sequencing methods, the clonal dynamics of initial engraftment and subsequent serial propagation of primary and metastatic human breast cancers in immunodeficient mice. In all 15 cases examined, clonal selection on engraftment was observed in both primary and metastatic breast tumours, varying in degree from extreme selective engraftment of minor (<5% of starting population) clones to moderate, polyclonal engraftment. Furthermore, ongoing clonal dynamics during serial passaging is a feature of tumours experiencing modest initial selection. Through single-cell sequencing, we show that major mutation clusters estimated from tumour population sequencing relate predictably to the most abundant clonal genotypes, even in clonally complex and rapidly evolving cases. Finally, we show that similar clonal expansion patterns can emerge in independent grafts of the same starting tumour population, indicating that genomic aberrations can be reproducible determinants of evolutionary trajectories. Our results show that measurement of genomically defined clonal population dynamics will be highly informative for functional studies using patient-derived breast cancer xenoengraftment.
Conflict of interest statement
[Competing Interests] The authors declare that they have no competing financial interests.
Figures




Comment in
-
Clonal evolution in cancer: a tale of twisted twines.Cell Stem Cell. 2015 Jan 8;16(1):11-2. doi: 10.1016/j.stem.2014.12.011. Cell Stem Cell. 2015. PMID: 25575078
Similar articles
-
The clonal and mutational evolution spectrum of primary triple-negative breast cancers.Nature. 2012 Apr 4;486(7403):395-9. doi: 10.1038/nature10933. Nature. 2012. PMID: 22495314 Free PMC article.
-
Clonal evolution in cancer: a tale of twisted twines.Cell Stem Cell. 2015 Jan 8;16(1):11-2. doi: 10.1016/j.stem.2014.12.011. Cell Stem Cell. 2015. PMID: 25575078
-
Breast tumours maintain a reservoir of subclonal diversity during expansion.Nature. 2021 Apr;592(7853):302-308. doi: 10.1038/s41586-021-03357-x. Epub 2021 Mar 24. Nature. 2021. PMID: 33762732 Free PMC article.
-
Clonal evolution in cancer.Nature. 2012 Jan 18;481(7381):306-13. doi: 10.1038/nature10762. Nature. 2012. PMID: 22258609 Free PMC article. Review.
-
Patient-derived xenograft models of breast cancer and their predictive power.Breast Cancer Res. 2015 Feb 10;17(1):17. doi: 10.1186/s13058-015-0523-1. Breast Cancer Res. 2015. PMID: 25849559 Free PMC article. Review.
Cited by
-
Laboratory Models for Investigating Breast Cancer Therapy Resistance and Metastasis.Front Oncol. 2021 Mar 10;11:645698. doi: 10.3389/fonc.2021.645698. eCollection 2021. Front Oncol. 2021. PMID: 33777805 Free PMC article. Review.
-
Linking primary and metastatic tumour re-initiation.Nat Cell Biol. 2015 May;17(5):542-3. doi: 10.1038/ncb3173. Nat Cell Biol. 2015. PMID: 25925584
-
The landscape of chromosomal aberrations in breast cancer mouse models reveals driver-specific routes to tumorigenesis.Nat Commun. 2016 Jul 4;7:12160. doi: 10.1038/ncomms12160. Nat Commun. 2016. PMID: 27374210 Free PMC article.
-
Mouse models of diffuse large B cell lymphoma.Front Immunol. 2023 Dec 6;14:1313371. doi: 10.3389/fimmu.2023.1313371. eCollection 2023. Front Immunol. 2023. PMID: 38124747 Free PMC article. Review.
-
Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery.Br J Cancer. 2020 Mar;122(6):735-744. doi: 10.1038/s41416-019-0672-6. Epub 2020 Jan 2. Br J Cancer. 2020. PMID: 31894140 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

