Towards a therapy for Angelman syndrome by targeting a long non-coding RNA
- PMID: 25470045
- PMCID: PMC4351819
- DOI: 10.1038/nature13975
Towards a therapy for Angelman syndrome by targeting a long non-coding RNA
Abstract
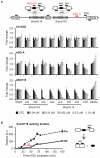
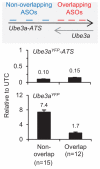
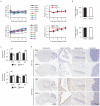
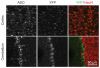
Angelman syndrome is a single-gene disorder characterized by intellectual disability, developmental delay, behavioural uniqueness, speech impairment, seizures and ataxia. It is caused by maternal deficiency of the imprinted gene UBE3A, encoding an E3 ubiquitin ligase. All patients carry at least one copy of paternal UBE3A, which is intact but silenced by a nuclear-localized long non-coding RNA, UBE3A antisense transcript (UBE3A-ATS). Murine Ube3a-ATS reduction by either transcription termination or topoisomerase I inhibition has been shown to increase paternal Ube3a expression. Despite a clear understanding of the disease-causing event in Angelman syndrome and the potential to harness the intact paternal allele to correct the disease, no gene-specific treatment exists for patients. Here we developed a potential therapeutic intervention for Angelman syndrome by reducing Ube3a-ATS with antisense oligonucleotides (ASOs). ASO treatment achieved specific reduction of Ube3a-ATS and sustained unsilencing of paternal Ube3a in neurons in vitro and in vivo. Partial restoration of UBE3A protein in an Angelman syndrome mouse model ameliorated some cognitive deficits associated with the disease. Although additional studies of phenotypic correction are needed, we have developed a sequence-specific and clinically feasible method to activate expression of the paternal Ube3a allele.
Figures



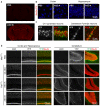
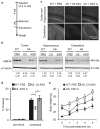
Comment in
-
Neurodevelopmental disorders. Unmuting Ube3a in mice alleviates Angelman syndrome.Nat Rev Neurol. 2015 Feb;11(2):66. doi: 10.1038/nrneurol.2014.240. Epub 2014 Dec 16. Nat Rev Neurol. 2015. PMID: 25511900 No abstract available.
Similar articles
-
Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model.PLoS Genet. 2013;9(12):e1004039. doi: 10.1371/journal.pgen.1004039. Epub 2013 Dec 26. PLoS Genet. 2013. PMID: 24385930 Free PMC article.
-
Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a.Hum Mol Genet. 2012 Jul 1;21(13):3001-12. doi: 10.1093/hmg/dds130. Epub 2012 Apr 5. Hum Mol Genet. 2012. PMID: 22493002 Free PMC article.
-
Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons.Nature. 2011 Dec 21;481(7380):185-9. doi: 10.1038/nature10726. Nature. 2011. PMID: 22190039 Free PMC article.
-
Gene Therapy for Angelman Syndrome: Contemporary Approaches and Future Endeavors.Curr Gene Ther. 2020;19(6):359-366. doi: 10.2174/1566523220666200107151025. Curr Gene Ther. 2020. PMID: 31914913 Review.
-
UBE3A reinstatement as a disease-modifying therapy for Angelman syndrome.Dev Med Child Neurol. 2021 Jul;63(7):802-807. doi: 10.1111/dmcn.14831. Epub 2021 Feb 4. Dev Med Child Neurol. 2021. PMID: 33543479 Free PMC article. Review.
Cited by
-
Identification of Natural Antisense Transcripts in Mouse Brain and Their Association With Autism Spectrum Disorder Risk Genes.Front Mol Neurosci. 2021 Feb 25;14:624881. doi: 10.3389/fnmol.2021.624881. eCollection 2021. Front Mol Neurosci. 2021. PMID: 33716665 Free PMC article.
-
The long non-coding RNA HLNC1 potentiates hepatocellular carcinoma progression via interaction with USP49.J Clin Lab Anal. 2020 Nov;34(11):e23462. doi: 10.1002/jcla.23462. Epub 2020 Jul 21. J Clin Lab Anal. 2020. PMID: 32691951 Free PMC article.
-
Upregulation of Haploinsufficient Gene Expression in the Brain by Targeting a Long Non-coding RNA Improves Seizure Phenotype in a Model of Dravet Syndrome.EBioMedicine. 2016 Jul;9:257-277. doi: 10.1016/j.ebiom.2016.05.011. Epub 2016 May 13. EBioMedicine. 2016. PMID: 27333023 Free PMC article.
-
RBFOX1 and RBFOX2 are dispensable in iPSCs and iPSC-derived neurons and do not contribute to neural-specific paternal UBE3A silencing.Sci Rep. 2016 May 5;6:25368. doi: 10.1038/srep25368. Sci Rep. 2016. PMID: 27146458 Free PMC article.
-
Angelman Syndrome.Neurotherapeutics. 2015 Jul;12(3):641-50. doi: 10.1007/s13311-015-0361-y. Neurotherapeutics. 2015. PMID: 26040994 Free PMC article. Review.
References
-
- Williams CA, Driscoll DJ, Dagli AI. Clinical and genetic aspects of Angelman syndrome. Genetics in medicine : official journal of the American College of Medical Genetics. 2010;12:385–395. - PubMed
-
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nature genetics. 1997;15:70–73. - PubMed
-
- Matsuura T, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nature genetics. 1997;15:74–77. - PubMed
-
- Albrecht U, et al. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nature genetics. 1997;17:75–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

