Haploinsufficiency of MYBPC3 exacerbates the development of hypertrophic cardiomyopathy in heterozygous mice
- PMID: 25463273
- PMCID: PMC4642280
- DOI: 10.1016/j.yjmcc.2014.11.018
Haploinsufficiency of MYBPC3 exacerbates the development of hypertrophic cardiomyopathy in heterozygous mice
Abstract
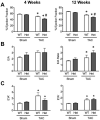
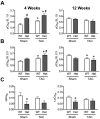
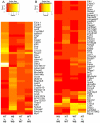
Mutations in MYBPC3, the gene encoding cardiac myosin binding protein-C (cMyBP-C), account for ~40% of hypertrophic cardiomyopathy (HCM) cases. Most pathological MYBPC3 mutations encode truncated protein products not found in tissue. Reduced protein levels occur in symptomatic heterozygous human HCM carriers, suggesting haploinsufficiency as an underlying mechanism of disease. However, we do not know if reduced cMyBP-C content results from, or initiates the development of HCM. In previous studies, heterozygous (HET) mice with a MYBPC3 C'-terminal truncation mutation and normal cMyBP-C levels show altered contractile function prior to any overt hypertrophy. Therefore, this study aimed to test whether haploinsufficiency occurs, with decreased cMyBP-C content, following cardiac stress and whether the functional impairment in HET MYBPC3 hearts leads to worsened disease progression. To address these questions, transverse aortic constriction (TAC) was performed on three-month-old wild-type (WT) and HET MYBPC3-truncation mutant mice and then characterized at 4 and 12weeks post-surgery. HET-TAC mice showed increased hypertrophy and reduced ejection fraction compared to WT-TAC mice. At 4weeks post-surgery, HET myofilaments showed significantly reduced cMyBP-C content. Functionally, HET-TAC cardiomyocytes showed impaired force generation, higher Ca(2+) sensitivity, and blunted length-dependent increase in force generation. RNA sequencing revealed several differentially regulated genes between HET and WT groups, including regulators of remodeling and hypertrophic response. Collectively, these results demonstrate that haploinsufficiency occurs in HET MYBPC3 mutant carriers following stress, causing, in turn, reduced cMyBP-C content and exacerbating the development of dysfunction at myofilament and whole-heart levels.
Keywords: Cardiac myosin binding protein-C; Haploinsufficiency; Hypertrophic cardiomyopathy; MYBPC3; Mouse models.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures


Comment in
-
Haploinsufficiency MYBPC3 mutations: another stress induced cardiomyopathy? Let's take a look!J Mol Cell Cardiol. 2015 Feb;79:284-6. doi: 10.1016/j.yjmcc.2014.12.008. Epub 2014 Dec 16. J Mol Cell Cardiol. 2015. PMID: 25524041 No abstract available.
Similar articles
-
Contractile dysfunction in a mouse model expressing a heterozygous MYBPC3 mutation associated with hypertrophic cardiomyopathy.Am J Physiol Heart Circ Physiol. 2014 Mar;306(6):H807-15. doi: 10.1152/ajpheart.00913.2013. Epub 2014 Jan 24. Am J Physiol Heart Circ Physiol. 2014. PMID: 24464755 Free PMC article.
-
Variable cardiac myosin binding protein-C expression in the myofilaments due to MYBPC3 mutations in hypertrophic cardiomyopathy.J Mol Cell Cardiol. 2018 Oct;123:59-63. doi: 10.1016/j.yjmcc.2018.08.023. Epub 2018 Aug 28. J Mol Cell Cardiol. 2018. PMID: 30170119
-
Contractile abnormalities and altered drug response in engineered heart tissue from Mybpc3-targeted knock-in mice.J Mol Cell Cardiol. 2013 Oct;63:189-98. doi: 10.1016/j.yjmcc.2013.07.011. Epub 2013 Jul 26. J Mol Cell Cardiol. 2013. PMID: 23896226
-
The mechanics of the heart: zooming in on hypertrophic cardiomyopathy and cMyBP-C.FEBS Lett. 2022 Mar;596(6):703-746. doi: 10.1002/1873-3468.14301. Epub 2022 Feb 28. FEBS Lett. 2022. PMID: 35224729 Review.
-
Allelic imbalance and haploinsufficiency in MYBPC3-linked hypertrophic cardiomyopathy.Pflugers Arch. 2019 May;471(5):781-793. doi: 10.1007/s00424-018-2226-9. Epub 2018 Nov 20. Pflugers Arch. 2019. PMID: 30456444 Free PMC article. Review.
Cited by
-
Alterations in Multi-Scale Cardiac Architecture in Association With Phosphorylation of Myosin Binding Protein-C.J Am Heart Assoc. 2016 Mar 15;5(3):e002836. doi: 10.1161/JAHA.115.002836. J Am Heart Assoc. 2016. PMID: 27068630 Free PMC article.
-
The genetic landscape of cardiomyopathy and its role in heart failure.Cell Metab. 2015 Feb 3;21(2):174-182. doi: 10.1016/j.cmet.2015.01.013. Cell Metab. 2015. PMID: 25651172 Free PMC article. Review.
-
Hnf1b haploinsufficiency differentially affects developmental target genes in a new renal cysts and diabetes mouse model.Dis Model Mech. 2021 May 1;14(5):dmm047498. doi: 10.1242/dmm.047498. Epub 2021 May 4. Dis Model Mech. 2021. PMID: 33737325 Free PMC article.
-
Proteolytic degradation of atrial sarcomere proteins underlies contractile defects in atrial fibrillation.Am J Physiol Heart Circ Physiol. 2024 Aug 1;327(2):H460-H472. doi: 10.1152/ajpheart.00148.2024. Epub 2024 Jun 28. Am J Physiol Heart Circ Physiol. 2024. PMID: 38940916
-
Thick Filament Protein Network, Functions, and Disease Association.Compr Physiol. 2018 Mar 13;8(2):631-709. doi: 10.1002/cphy.c170023. Compr Physiol. 2018. PMID: 29687901 Free PMC article. Review.
References
-
- Maron B, Gardin J, Flack J, Gidding S, Kurosaki T, Bild D. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: echocardiographic analysis of 4111 subjects in the CARDIA study. Circulation. 1995;92:785–9. - PubMed
-
- Watkins H, Conner D, Thierfelder L, Jarcho JA, MacRae C, McKenna WJ, et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat Genet. 1995;11:434–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

