CREB regulates memory allocation in the insular cortex
- PMID: 25454591
- PMCID: PMC4743759
- DOI: 10.1016/j.cub.2014.10.018
CREB regulates memory allocation in the insular cortex
Abstract
The molecular and cellular mechanisms of memory storage have attracted a great deal of attention. By comparison, little is known about memory allocation, the process that determines which specific neurons in a neural network will store a given memory. Previous studies demonstrated that memory allocation is not random in the amygdala; these studies showed that amygdala neurons with higher levels of the cyclic-AMP-response-element-binding protein (CREB) are more likely to be recruited into encoding and storing fear memory. To determine whether specific mechanisms also regulate memory allocation in other brain regions and whether CREB also has a role in this process, we studied insular cortical memory representations for conditioned taste aversion (CTA). In this task, an animal learns to associate a taste (conditioned stimulus [CS]) with the experience of malaise (such as that induced by LiCl; unconditioned stimulus [US]). The insular cortex is required for CTA memory formation and retrieval. CTA learning activates a subpopulation of neurons in this structure, and the insular cortex and the basolateral amygdala (BLA) interact during CTA formation. Here, we used a combination of approaches, including viral vector transfections of insular cortex, arc fluorescence in situ hybridization (FISH), and designer receptors exclusively activated by designer drugs (DREADD) system, to show that CREB levels determine which insular cortical neurons go on to encode a given conditioned taste memory.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures
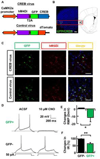
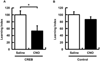
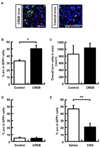
Similar articles
-
Cortico-amygdala interaction determines the insular cortical neurons involved in taste memory retrieval.Mol Brain. 2020 Jul 28;13(1):107. doi: 10.1186/s13041-020-00646-w. Mol Brain. 2020. PMID: 32723372 Free PMC article.
-
cAMP response element-binding protein in the amygdala is required for long- but not short-term conditioned taste aversion memory.J Neurosci. 1997 Nov 1;17(21):8443-50. doi: 10.1523/JNEUROSCI.17-21-08443.1997. J Neurosci. 1997. PMID: 9334416 Free PMC article.
-
Activity of Insula to Basolateral Amygdala Projecting Neurons is Necessary and Sufficient for Taste Valence Representation.J Neurosci. 2019 Nov 20;39(47):9369-9382. doi: 10.1523/JNEUROSCI.0752-19.2019. Epub 2019 Oct 9. J Neurosci. 2019. PMID: 31597726 Free PMC article.
-
Interplay of amygdala and insular cortex during and after associative taste aversion memory formation.Rev Neurosci. 2012;23(5-6):463-71. doi: 10.1515/revneuro-2012-0056. Rev Neurosci. 2012. PMID: 23001315 Review.
-
Inducible repression of CREB function disrupts amygdala-dependent memory.Neurobiol Learn Mem. 2004 Sep;82(2):159-63. doi: 10.1016/j.nlm.2004.05.008. Neurobiol Learn Mem. 2004. PMID: 15341801 Review.
Cited by
-
A locus coeruleus-dorsal CA1 dopaminergic circuit modulates memory linking.Neuron. 2022 Oct 19;110(20):3374-3388.e8. doi: 10.1016/j.neuron.2022.08.001. Epub 2022 Aug 29. Neuron. 2022. PMID: 36041433 Free PMC article.
-
DREADDS: Use and application in behavioral neuroscience.Behav Neurosci. 2016 Apr;130(2):137-55. doi: 10.1037/bne0000135. Epub 2016 Feb 25. Behav Neurosci. 2016. PMID: 26913540 Free PMC article. Review.
-
Growth hormone biases amygdala network activation after fear learning.Transl Psychiatry. 2016 Nov 29;6(11):e960. doi: 10.1038/tp.2016.203. Transl Psychiatry. 2016. PMID: 27898076 Free PMC article.
-
The role of intrinsic excitability in the evolution of memory: Significance in memory allocation, consolidation, and updating.Neurobiol Learn Mem. 2020 Sep;173:107266. doi: 10.1016/j.nlm.2020.107266. Epub 2020 Jun 5. Neurobiol Learn Mem. 2020. PMID: 32512183 Free PMC article. Review.
-
Opto-extinction of a threat memory in mice.Brain Res Bull. 2022 Dec;191:61-68. doi: 10.1016/j.brainresbull.2022.10.012. Epub 2022 Oct 21. Brain Res Bull. 2022. PMID: 36279984 Free PMC article.
References
-
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, Josselyn SA. Neuronal competition and selection during memory formation. Science. 2007;316:457–460. - PubMed
-
- Han JH, Kushner SA, Yiu AP, Hsiang HL, Buch T, Waisman A, Bontempi B, Neve RL, Frankland PW, Josselyn SA. Selective erasure of a fear memory. Science. 2009;323:1492–1496. - PubMed
-
- Kim J, Kwon JT, Kim HS, Josselyn SA, Han JH. Memory recall and modifications by activating neurons with elevated CREB. Nature neuroscience. 2014;17:65–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

