Closing the cohesin ring: structure and function of its Smc3-kleisin interface
- PMID: 25414305
- PMCID: PMC4300515
- DOI: 10.1126/science.1256917
Closing the cohesin ring: structure and function of its Smc3-kleisin interface
Abstract
Through their association with a kleisin subunit (Scc1), cohesin's Smc1 and Smc3 subunits are thought to form tripartite rings that mediate sister chromatid cohesion. Unlike the structure of Smc1/Smc3 and Smc1/Scc1 interfaces, that of Smc3/Scc1 is not known. Disconnection of this interface is thought to release cohesin from chromosomes in a process regulated by acetylation. We show here that the N-terminal domain of yeast Scc1 contains two α helices, forming a four-helix bundle with the coiled coil emerging from Smc3's adenosine triphosphatase head. Mutations affecting this interaction compromise cohesin's association with chromosomes. The interface is far from Smc3 residues, whose acetylation prevents cohesin's dissociation from chromosomes. Cohesin complexes holding chromatids together in vivo do indeed have the configuration of hetero-trimeric rings, and sister DNAs are entrapped within these.
Copyright © 2014, American Association for the Advancement of Science.
Figures
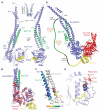

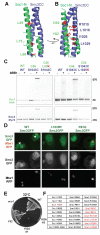
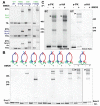
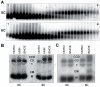
Comment in
-
Cell biology: cohesin rings leave loose ends.Curr Biol. 2015 Feb 2;25(3):R108-R110. doi: 10.1016/j.cub.2014.12.015. Curr Biol. 2015. PMID: 25649818 Free PMC article.
Similar articles
-
Releasing Activity Disengages Cohesin's Smc3/Scc1 Interface in a Process Blocked by Acetylation.Mol Cell. 2016 Feb 18;61(4):563-574. doi: 10.1016/j.molcel.2016.01.026. Mol Cell. 2016. PMID: 26895425 Free PMC article.
-
Sister DNA Entrapment between Juxtaposed Smc Heads and Kleisin of the Cohesin Complex.Mol Cell. 2019 Jul 25;75(2):224-237.e5. doi: 10.1016/j.molcel.2019.05.023. Epub 2019 Jun 11. Mol Cell. 2019. PMID: 31201089 Free PMC article.
-
The structure of the cohesin ATPase elucidates the mechanism of SMC-kleisin ring opening.Nat Struct Mol Biol. 2020 Mar;27(3):233-239. doi: 10.1038/s41594-020-0379-7. Epub 2020 Feb 17. Nat Struct Mol Biol. 2020. PMID: 32066964 Free PMC article.
-
The torments of the cohesin ring.Nucleus. 2017 May 4;8(3):261-267. doi: 10.1080/19491034.2017.1295200. Epub 2017 Feb 27. Nucleus. 2017. PMID: 28453390 Free PMC article. Review.
-
The structure and function of SMC and kleisin complexes.Annu Rev Biochem. 2005;74:595-648. doi: 10.1146/annurev.biochem.74.082803.133219. Annu Rev Biochem. 2005. PMID: 15952899 Review.
Cited by
-
Chromosome structure in Drosophila is determined by boundary pairing not loop extrusion.Elife. 2024 Aug 7;13:RP94070. doi: 10.7554/eLife.94070. Elife. 2024. PMID: 39110499 Free PMC article.
-
Cell biology: cohesin rings leave loose ends.Curr Biol. 2015 Feb 2;25(3):R108-R110. doi: 10.1016/j.cub.2014.12.015. Curr Biol. 2015. PMID: 25649818 Free PMC article.
-
The CDK Pef1 and protein phosphatase 4 oppose each other for regulating cohesin binding to fission yeast chromosomes.Elife. 2020 Jan 2;9:e50556. doi: 10.7554/eLife.50556. Elife. 2020. PMID: 31895039 Free PMC article.
-
Towards a Unified Model of SMC Complex Function.Curr Biol. 2018 Nov 5;28(21):R1266-R1281. doi: 10.1016/j.cub.2018.08.034. Curr Biol. 2018. PMID: 30399354 Free PMC article. Review.
-
Mediator recruits the cohesin loader Scc2 to RNA Pol II-transcribed genes and promotes sister chromatid cohesion.Curr Biol. 2022 Jul 11;32(13):2884-2896.e6. doi: 10.1016/j.cub.2022.05.019. Epub 2022 Jun 1. Curr Biol. 2022. PMID: 35654035 Free PMC article.
References
-
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. - PubMed
-
- Nasmyth K. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol. 2011;13:1170–1177. - PubMed
-
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. - PubMed
-
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular Architecture of SMC Proteins and the Yeast Cohesin Complex. Mol Cell. 2002;9:773–788. - PubMed
-
- Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, Nasmyth K. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

