Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions
- PMID: 25412268
- PMCID: PMC4239116
- DOI: 10.1371/journal.ppat.1004512
Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions
Abstract
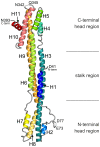
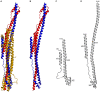
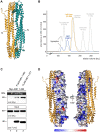
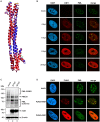
PML nuclear bodies (PML-NBs) are enigmatic structures of the cell nucleus that act as key mediators of intrinsic immunity against viral pathogens. PML itself is a member of the E3-ligase TRIM family of proteins that regulates a variety of innate immune signaling pathways. Consequently, viruses have evolved effector proteins to modify PML-NBs; however, little is known concerning structure-function relationships of viral antagonists. The herpesvirus human cytomegalovirus (HCMV) expresses the abundant immediate-early protein IE1 that colocalizes with PML-NBs and induces their dispersal, which correlates with the antagonization of NB-mediated intrinsic immunity. Here, we delineate the molecular basis for this antagonization by presenting the first crystal structure for the evolutionary conserved primate cytomegalovirus IE1 proteins. We show that IE1 consists of a globular core (IE1CORE) flanked by intrinsically disordered regions. The 2.3 Å crystal structure of IE1CORE displays an all α-helical, femur-shaped fold, which lacks overall fold similarity with known protein structures, but shares secondary structure features recently observed in the coiled-coil domain of TRIM proteins. Yeast two-hybrid and coimmunoprecipitation experiments demonstrate that IE1CORE binds efficiently to the TRIM family member PML, and is able to induce PML deSUMOylation. Intriguingly, this results in the release of NB-associated proteins into the nucleoplasm, but not of PML itself. Importantly, we show that PML deSUMOylation by IE1CORE is sufficient to antagonize PML-NB-instituted intrinsic immunity. Moreover, co-immunoprecipitation experiments demonstrate that IE1CORE binds via the coiled-coil domain to PML and also interacts with TRIM5α We propose that IE1CORE sequesters PML and possibly other TRIM family members via structural mimicry using an extended binding surface formed by the coiled-coil region. This mode of interaction might render the antagonizing activity less susceptible to mutational escape.
Conflict of interest statement
The authors have declared that no competing intests exist.
Figures


Similar articles
-
Emerging Role of PML Nuclear Bodies in Innate Immune Signaling.J Virol. 2016 Jun 10;90(13):5850-5854. doi: 10.1128/JVI.01979-15. Print 2016 Jul 1. J Virol. 2016. PMID: 27053550 Free PMC article. Review.
-
Characterization of Recombinant Human Cytomegaloviruses Encoding IE1 Mutants L174P and 1-382 Reveals that Viral Targeting of PML Bodies Perturbs both Intrinsic and Innate Immune Responses.J Virol. 2015 Nov 11;90(3):1190-205. doi: 10.1128/JVI.01973-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26559840 Free PMC article.
-
Cytomegalovirus immediate-early 1 proteins form a structurally distinct protein class with adaptations determining cross-species barriers.PLoS Pathog. 2021 Aug 9;17(8):e1009863. doi: 10.1371/journal.ppat.1009863. eCollection 2021 Aug. PLoS Pathog. 2021. PMID: 34370791 Free PMC article.
-
The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation.J Virol. 2017 Jan 31;91(4):e02049-16. doi: 10.1128/JVI.02049-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27903803 Free PMC article.
-
The Human CMV IE1 Protein: An Offender of PML Nuclear Bodies.Adv Anat Embryol Cell Biol. 2017;223:77-94. doi: 10.1007/978-3-319-53168-7_4. Adv Anat Embryol Cell Biol. 2017. PMID: 28528440 Review.
Cited by
-
Emerging Role of PML Nuclear Bodies in Innate Immune Signaling.J Virol. 2016 Jun 10;90(13):5850-5854. doi: 10.1128/JVI.01979-15. Print 2016 Jul 1. J Virol. 2016. PMID: 27053550 Free PMC article. Review.
-
Human Cytomegalovirus Immediate-Early 1 Protein Rewires Upstream STAT3 to Downstream STAT1 Signaling Switching an IL6-Type to an IFNγ-Like Response.PLoS Pathog. 2016 Jul 7;12(7):e1005748. doi: 10.1371/journal.ppat.1005748. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27387064 Free PMC article.
-
Regulation of Tripartite Motif-Containing Proteins on Immune Response and Viral Evasion.Front Microbiol. 2021 Dec 1;12:794882. doi: 10.3389/fmicb.2021.794882. eCollection 2021. Front Microbiol. 2021. PMID: 34925304 Free PMC article. Review.
-
Dual functions of Aire CARD multimerization in the transcriptional regulation of T cell tolerance.Nat Commun. 2020 Apr 2;11(1):1625. doi: 10.1038/s41467-020-15448-w. Nat Commun. 2020. PMID: 32242017 Free PMC article.
-
Expression of Human Cytomegalovirus IE1 Leads to Accumulation of Mono-SUMOylated PML That Is Protected from Degradation by Herpes Simplex Virus 1 ICP0.J Virol. 2018 Nov 12;92(23):e01452-18. doi: 10.1128/JVI.01452-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30258013 Free PMC article.
References
-
- Jensen K, Shiels C, Freemont PS (2001) PML protein isoforms and the RBCC/TRIM motif. Oncogene 20: 7223–7233. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

