RNA regulons in Hox 5' UTRs confer ribosome specificity to gene regulation
- PMID: 25409156
- PMCID: PMC4353651
- DOI: 10.1038/nature14010
RNA regulons in Hox 5' UTRs confer ribosome specificity to gene regulation
Abstract
Emerging evidence suggests that the ribosome has a regulatory function in directing how the genome is translated in time and space. However, how this regulation is encoded in the messenger RNA sequence remains largely unknown. Here we uncover unique RNA regulons embedded in homeobox (Hox) 5' untranslated regions (UTRs) that confer ribosome-mediated control of gene expression. These structured RNA elements, resembling viral internal ribosome entry sites (IRESs), are found in subsets of Hox mRNAs. They facilitate ribosome recruitment and require the ribosomal protein RPL38 for their activity. Despite numerous layers of Hox gene regulation, these IRES elements are essential for converting Hox transcripts into proteins to pattern the mammalian body plan. This specialized mode of IRES-dependent translation is enabled by an additional regulatory element that we term the translation inhibitory element (TIE), which blocks cap-dependent translation of transcripts. Together, these data uncover a new paradigm for ribosome-mediated control of gene expression and organismal development.
Conflict of interest statement
The authors declare no competing interests.
Figures
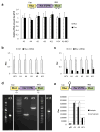


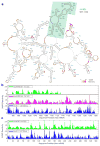

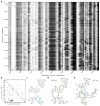

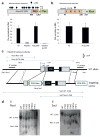




Comment in
-
Molecular biology: Entry signals control development.Nature. 2015 Jan 1;517(7532):24-5. doi: 10.1038/nature14069. Epub 2014 Nov 19. Nature. 2015. PMID: 25409148 Free PMC article.
Similar articles
-
Cis-regulatory RNA elements that regulate specialized ribosome activity.RNA Biol. 2015;12(10):1083-7. doi: 10.1080/15476286.2015.1085149. Epub 2015 Sep 1. RNA Biol. 2015. PMID: 26327194 Free PMC article. Review.
-
Gene- and Species-Specific Hox mRNA Translation by Ribosome Expansion Segments.Mol Cell. 2020 Dec 17;80(6):980-995.e13. doi: 10.1016/j.molcel.2020.10.023. Epub 2020 Nov 16. Mol Cell. 2020. PMID: 33202249 Free PMC article.
-
Molecular biology: Entry signals control development.Nature. 2015 Jan 1;517(7532):24-5. doi: 10.1038/nature14069. Epub 2014 Nov 19. Nature. 2015. PMID: 25409148 Free PMC article.
-
A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry.Mol Cell Biol. 2005 Sep;25(17):7879-88. doi: 10.1128/MCB.25.17.7879-7888.2005. Mol Cell Biol. 2005. PMID: 16107731 Free PMC article.
-
Investigation of interactions of polypyrimidine tract-binding protein with artificial internal ribosome entry segments.Biochem Soc Trans. 2005 Dec;33(Pt 6):1483-6. doi: 10.1042/BST0331483. Biochem Soc Trans. 2005. PMID: 16246151 Review.
Cited by
-
False-positive IRESes from Hoxa9 and other genes resulting from errors in mammalian 5' UTR annotations.Proc Natl Acad Sci U S A. 2022 Sep 6;119(36):e2122170119. doi: 10.1073/pnas.2122170119. Epub 2022 Aug 29. Proc Natl Acad Sci U S A. 2022. PMID: 36037358 Free PMC article.
-
Are there roles for heterogeneous ribosomes during sleep in the rodent brain?Front Mol Biosci. 2022 Oct 6;9:1008921. doi: 10.3389/fmolb.2022.1008921. eCollection 2022. Front Mol Biosci. 2022. PMID: 36275625 Free PMC article.
-
Designing synthetic RNAs to determine the relevance of structural motifs in picornavirus IRES elements.Sci Rep. 2016 Apr 7;6:24243. doi: 10.1038/srep24243. Sci Rep. 2016. PMID: 27053355 Free PMC article.
-
Analysis of the 5' Untranslated Region Length-Dependent Control of Gene Expression in Maize: A Case Study with the ZmLAZ1 Gene Family.Genes (Basel). 2024 Jul 29;15(8):994. doi: 10.3390/genes15080994. Genes (Basel). 2024. PMID: 39202355 Free PMC article.
-
The m6A epitranscriptome: transcriptome plasticity in brain development and function.Nat Rev Neurosci. 2020 Jan;21(1):36-51. doi: 10.1038/s41583-019-0244-z. Epub 2019 Dec 5. Nat Rev Neurosci. 2020. PMID: 31804615 Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

