Circular RNAs: diversity of form and function
- PMID: 25404635
- PMCID: PMC4238349
- DOI: 10.1261/rna.047126.114
Circular RNAs: diversity of form and function
Abstract
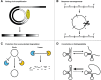
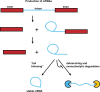
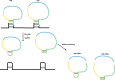
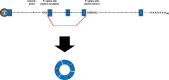
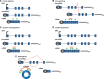
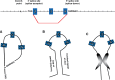
It is now clear that there is a diversity of circular RNAs in biological systems. Circular RNAs can be produced by the direct ligation of 5' and 3' ends of linear RNAs, as intermediates in RNA processing reactions, or by "backsplicing," wherein a downstream 5' splice site (splice donor) is joined to an upstream 3' splice site (splice acceptor). Circular RNAs have unique properties including the potential for rolling circle amplification of RNA, the ability to rearrange the order of genomic information, protection from exonucleases, and constraints on RNA folding. Circular RNAs can function as templates for viroid and viral replication, as intermediates in RNA processing reactions, as regulators of transcription in cis, as snoRNAs, and as miRNA sponges. Herein, we review the breadth of circular RNAs, their biogenesis and metabolism, and their known and anticipated functions.
Keywords: backsplicing; circRNA; circular RNA; intermediates; intron; nonlinear; splicing.
© 2014 Lasda and Parker; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures
Similar articles
-
Circular RNA Splicing.Adv Exp Med Biol. 2018;1087:41-52. doi: 10.1007/978-981-13-1426-1_4. Adv Exp Med Biol. 2018. PMID: 30259356 Review.
-
Circular RNAs: Identification, biogenesis and function.Biochim Biophys Acta. 2016 Jan;1859(1):163-8. doi: 10.1016/j.bbagrm.2015.07.007. Epub 2015 Jul 11. Biochim Biophys Acta. 2016. PMID: 26171810 Review.
-
Circular RNAs: relics of precellular evolution?Proc Natl Acad Sci U S A. 1989 Dec;86(23):9370-4. doi: 10.1073/pnas.86.23.9370. Proc Natl Acad Sci U S A. 1989. PMID: 2480600 Free PMC article.
-
Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis delta virus: variations on a theme.RNA Biol. 2011 Mar-Apr;8(2):200-6. doi: 10.4161/rna.8.2.14238. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21358283 Review.
-
Circular RNAs: Unexpected outputs of many protein-coding genes.RNA Biol. 2017 Aug 3;14(8):1007-1017. doi: 10.1080/15476286.2016.1227905. Epub 2016 Aug 29. RNA Biol. 2017. PMID: 27571848 Free PMC article. Review.
Cited by
-
Downregulation of hsa_circ_0006220 and its correlation with clinicopathological factors in human breast cancer.Gland Surg. 2021 Feb;10(2):816-825. doi: 10.21037/gs-21-42. Gland Surg. 2021. PMID: 33708563 Free PMC article.
-
Native Circular RNA Pulldown Method to Simultaneously Profile RNA and Protein Interactions.Methods Mol Biol. 2024;2765:299-309. doi: 10.1007/978-1-0716-3678-7_16. Methods Mol Biol. 2024. PMID: 38381346
-
ZEB1-Mediated Transcriptional Upregulation of circWWC3 Promotes Breast Cancer Progression through Activating Ras Signaling Pathway.Mol Ther Nucleic Acids. 2020 Aug 19;22:124-137. doi: 10.1016/j.omtn.2020.08.015. Online ahead of print. Mol Ther Nucleic Acids. 2020. PMID: 32916598 Free PMC article.
-
Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs.Nat Commun. 2016 Apr 6;7:11215. doi: 10.1038/ncomms11215. Nat Commun. 2016. PMID: 27050392 Free PMC article.
-
A systematic analysis of circRNAs in subnuclear compartments.RNA Biol. 2024 Jan;21(1):1-16. doi: 10.1080/15476286.2024.2395718. Epub 2024 Sep 10. RNA Biol. 2024. PMID: 39257052 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
