MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL
- PMID: 25403230
- PMCID: PMC4238046
- DOI: 10.1038/ncomms6413
MNKs act as a regulatory switch for eIF4E1 and eIF4E3 driven mRNA translation in DLBCL
Abstract
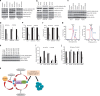
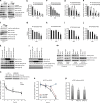
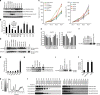
The phosphorylation of eIF4E1 at serine 209 by MNK1 or MNK2 has been shown to initiate oncogenic mRNA translation, a process that favours cancer development and maintenance. Here, we interrogate the MNK-eIF4E axis in diffuse large B-cell lymphoma (DLBCL) and show a distinct distribution of MNK1 and MNK2 in germinal centre B-cell (GCB) and activated B-cell (ABC) DLBCL. Despite displaying a differential distribution in GCB and ABC, both MNKs functionally complement each other to sustain cell survival. MNK inhibition ablates eIF4E1 phosphorylation and concurrently enhances eIF4E3 expression. Loss of MNK protein itself downregulates total eIF4E1 protein level by reducing eIF4E1 mRNA polysomal loading without affecting total mRNA level or stability. Enhanced eIF4E3 expression marginally suppresses eIF4E1-driven translation but exhibits a unique translatome that unveils a novel role for eIF4E3 in translation initiation. We propose that MNKs can modulate oncogenic translation by regulating eIF4E1-eIF4E3 levels and activity in DLBCL.
Figures

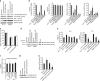


Similar articles
-
Distinct recruitment of human eIF4E isoforms to processing bodies and stress granules.BMC Mol Biol. 2016 Aug 30;17(1):21. doi: 10.1186/s12867-016-0072-x. BMC Mol Biol. 2016. PMID: 27578149 Free PMC article.
-
Major splice variants and multiple polyadenylation site utilization in mRNAs encoding human translation initiation factors eIF4E1 and eIF4E3 regulate the translational regulators?Mol Genet Genomics. 2018 Feb;293(1):167-186. doi: 10.1007/s00438-017-1375-4. Epub 2017 Sep 23. Mol Genet Genomics. 2018. PMID: 28942592
-
The MAP kinase-interacting kinases regulate cell migration, vimentin expression and eIF4E/CYFIP1 binding.Biochem J. 2015 Apr 1;467(1):63-76. doi: 10.1042/BJ20141066. Biochem J. 2015. PMID: 25588502
-
Mnks, eIF4E phosphorylation and cancer.Biochim Biophys Acta. 2015 Jul;1849(7):766-73. doi: 10.1016/j.bbagrm.2014.10.003. Epub 2014 Oct 23. Biochim Biophys Acta. 2015. PMID: 25450520 Review.
-
Progress in developing MNK inhibitors.Eur J Med Chem. 2021 Jul 5;219:113420. doi: 10.1016/j.ejmech.2021.113420. Epub 2021 Apr 2. Eur J Med Chem. 2021. PMID: 33892273 Review.
Cited by
-
Expression Profile and Prognostic Significance of Pivotal Regulators for N7-Methylguanosine Methylation in Diffuse Large B-Cell Lymphoma.Mol Biotechnol. 2024 Oct 22. doi: 10.1007/s12033-024-01264-w. Online ahead of print. Mol Biotechnol. 2024. PMID: 39436635
-
A network of RNA-binding proteins controls translation efficiency to activate anaerobic metabolism.Nat Commun. 2020 May 29;11(1):2677. doi: 10.1038/s41467-020-16504-1. Nat Commun. 2020. PMID: 32472050 Free PMC article.
-
RNA m6A methylation regulates dissemination of cancer cells by modulating expression and membrane localization of β-catenin.Mol Ther. 2022 Apr 6;30(4):1578-1596. doi: 10.1016/j.ymthe.2022.01.019. Epub 2022 Jan 14. Mol Ther. 2022. PMID: 35033632 Free PMC article.
-
eIF4E3 forms an active eIF4F complex during stresses (eIF4FS) targeting mTOR and re-programs the translatome.Nucleic Acids Res. 2021 May 21;49(9):5159-5176. doi: 10.1093/nar/gkab267. Nucleic Acids Res. 2021. PMID: 33893802 Free PMC article.
-
RNA-tethering assay and eIF4G:eIF4A obligate dimer design uncovers multiple eIF4F functional complexes.Nucleic Acids Res. 2020 Sep 4;48(15):8562-8575. doi: 10.1093/nar/gkaa646. Nucleic Acids Res. 2020. PMID: 32749456 Free PMC article.
References
-
- Ruggero D. et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-myc in lymphomagenesis. Nat. Med. 10, 484–486 (2004). - PubMed
-
- Gingras A. C., Raught B. & Sonenberg N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 (1999). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

