Alternative Pathway for the Reaction Catalyzed by DNA Dealkylase AlkB from Ab Initio QM/MM Calculations
- PMID: 25400523
- PMCID: PMC4230374
- DOI: 10.1021/ct500572t
Alternative Pathway for the Reaction Catalyzed by DNA Dealkylase AlkB from Ab Initio QM/MM Calculations
Abstract
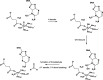
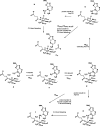
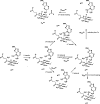
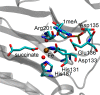
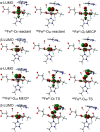
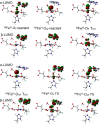
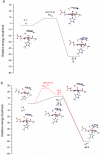
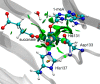
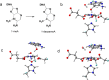
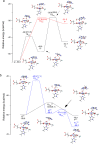
AlkB is the title enzyme of a family of DNA dealkylases that catalyze the direct oxidative dealkylation of nucleobases. The conventional mechanism for the dealkylation of N1-methyl adenine (1-meA) catalyzed by AlkB after the formation of FeIV-oxo is comprised by a reorientation of the oxo moiety, hydrogen abstraction, OH rebound from the Fe atom to the methyl adduct, and the dissociation of the resulting methoxide to obtain the repaired adenine base and formaldehyde. An alternative pathway with hydroxide as a ligand bound to the iron atom is proposed and investigated by QM/MM simulations. The results show OH- has a small impact on the barriers for the hydrogen abstraction and OH rebound steps. The effects of the enzyme and the OH- ligand on the hydrogen abstraction by the FeIV-oxo moiety are discussed in detail. The new OH rebound step is coupled with a proton transfer to the OH- ligand and results in a novel zwitterion intermediate. This zwitterion structure can also be characterized as Fe-O-C complex and facilitates the formation of formaldehyde. In contrast, for the pathway with H2O bound to iron, the hydroxyl product of the OH rebound step first needs to unbind from the metal center before transferring a proton to Glu136 or other residue/substrate. The consistency between our theoretical results and experimental findings is discussed. This study provides new insights into the oxidative repair mechanism of DNA repair by nonheme FeII and α-ketoglutarate (α-KG) dependent dioxygenases and a possible explanation for the substrate preference of AlkB.
Figures


Similar articles
-
Insights into the Direct Oxidative Repair of Etheno Lesions: MD and QM/MM Study on the Substrate Scope of ALKBH2 and AlkB.DNA Repair (Amst). 2020 Dec;96:102944. doi: 10.1016/j.dnarep.2020.102944. Epub 2020 Sep 9. DNA Repair (Amst). 2020. PMID: 33161373 Free PMC article.
-
Ab initio QM/MM calculations show an intersystem crossing in the hydrogen abstraction step in dealkylation catalyzed by AlkB.J Phys Chem B. 2013 May 30;117(21):6410-20. doi: 10.1021/jp403116e. Epub 2013 May 16. J Phys Chem B. 2013. PMID: 23642148
-
DFT study of a model system for the dealkylation step catalyzed by AlkB.Interdiscip Sci. 2010 Mar;2(1):70-7. doi: 10.1007/s12539-010-0092-z. Epub 2010 Jan 28. Interdiscip Sci. 2010. PMID: 20640798
-
The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond.J Biol Chem. 2015 Aug 21;290(34):20734-20742. doi: 10.1074/jbc.R115.656462. Epub 2015 Jul 7. J Biol Chem. 2015. PMID: 26152727 Free PMC article. Review.
-
Mechanistic Insight on the Activity and Substrate Selectivity of Nonheme Iron Dioxygenases.Chem Rec. 2018 Oct;18(10):1501-1516. doi: 10.1002/tcr.201800033. Epub 2018 Jun 7. Chem Rec. 2018. PMID: 29878456 Review.
Cited by
-
Protein effects in non-heme iron enzyme catalysis: insights from multiscale models.J Biol Inorg Chem. 2016 Sep;21(5-6):645-57. doi: 10.1007/s00775-016-1374-7. Epub 2016 Jun 30. J Biol Inorg Chem. 2016. PMID: 27364958 Review.
-
Computational Characterization of the Inhibition Mechanism of Xanthine Oxidoreductase by Topiroxostat.ACS Catal. 2023 May 5;13(9):6023-6043. doi: 10.1021/acscatal.3c01245. Epub 2023 Apr 18. ACS Catal. 2023. PMID: 37547543 Free PMC article.
-
Role of Structural Dynamics in Selectivity and Mechanism of Non-heme Fe(II) and 2-Oxoglutarate-Dependent Oxygenases Involved in DNA Repair.ACS Cent Sci. 2020 May 27;6(5):795-814. doi: 10.1021/acscentsci.0c00312. Epub 2020 May 8. ACS Cent Sci. 2020. PMID: 32490196 Free PMC article.
-
Insight into wild-type and T1372E TET2-mediated 5hmC oxidation using ab initio QM/MM calculations.Chem Sci. 2018 Sep 11;9(44):8433-8445. doi: 10.1039/c8sc02961j. eCollection 2018 Nov 28. Chem Sci. 2018. PMID: 30542593 Free PMC article.
-
Comparison of DNA and RNA substrate effects on TET2 structure.Adv Protein Chem Struct Biol. 2019;117:91-112. doi: 10.1016/bs.apcsb.2019.05.002. Epub 2019 Jun 11. Adv Protein Chem Struct Biol. 2019. PMID: 31564308 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
