PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript
- PMID: 25382312
- PMCID: PMC4228382
- DOI: 10.1038/ncomms6362
PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript
Abstract
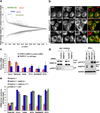
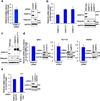
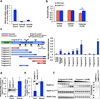
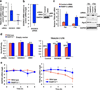
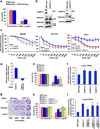
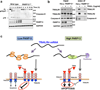
Poly(ADP-ribose) polymerase-13 (PARP13/ZAP/ZC3HAV1) is an antiviral factor, active against specific RNA viruses such as murine leukaemia virus, Sindbis virus and human immunodeficiency virus. During infection, PARP13 binds viral RNA via its four CCCH-type zinc-finger domains and targets it for degradation by recruiting cellular messenger RNA (mRNA) decay factors such as the exosome complex and XRN1. Here we show that PARP13 binds to and regulates cellular mRNAs in the absence of viral infection. Knockdown of PARP13 results in the misregulation of hundreds of transcripts. Among the most upregulated transcripts is TRAILR4 that encodes a decoy receptor for TRAIL-a pro-apoptotic cytokine that is a promising target for the therapeutic inhibition of cancers. PARP13 destabilizes TRAILR4 mRNA post-transcriptionally in an exosome-dependent manner by binding to a region in its 3' untranslated region. As a consequence, PARP13 represses TRAILR4 expression and increases cell sensitivity to TRAIL-mediated apoptosis, acting as a key regulator of the cellular response to TRAIL.
Figures


Similar articles
-
Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer.Trends Mol Med. 2015 Jun;21(6):373-84. doi: 10.1016/j.molmed.2015.03.002. Epub 2015 Apr 4. Trends Mol Med. 2015. PMID: 25851173 Free PMC article. Review.
-
Crystal structures and functional analysis of the ZnF5-WWE1-WWE2 region of PARP13/ZAP define a distinctive mode of engaging poly(ADP-ribose).Cell Rep. 2022 Oct 25;41(4):111529. doi: 10.1016/j.celrep.2022.111529. Cell Rep. 2022. PMID: 36288691 Free PMC article.
-
Zinc-finger antiviral protein acts as a tumor suppressor in colorectal cancer.Oncogene. 2020 Sep;39(37):5995-6008. doi: 10.1038/s41388-020-01416-7. Epub 2020 Aug 7. Oncogene. 2020. PMID: 32770142
-
Poly(ADP-ribose) potentiates ZAP antiviral activity.PLoS Pathog. 2022 Feb 7;18(2):e1009202. doi: 10.1371/journal.ppat.1009202. eCollection 2022 Feb. PLoS Pathog. 2022. PMID: 35130321 Free PMC article.
-
Versatility of the Zinc-Finger Antiviral Protein (ZAP) As a Modulator of Viral Infections.Int J Biol Sci. 2024 Aug 26;20(12):4585-4600. doi: 10.7150/ijbs.98029. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309436 Free PMC article. Review.
Cited by
-
LINE-1 Retroelements Get ZAPped!PLoS Genet. 2015 Jul 16;11(7):e1005364. doi: 10.1371/journal.pgen.1005364. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26182081 Free PMC article. No abstract available.
-
ZAP isoforms regulate unfolded protein response and epithelial- mesenchymal transition.Proc Natl Acad Sci U S A. 2022 Aug 2;119(31):e2121453119. doi: 10.1073/pnas.2121453119. Epub 2022 Jul 26. Proc Natl Acad Sci U S A. 2022. PMID: 35881805 Free PMC article.
-
Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer.Trends Mol Med. 2015 Jun;21(6):373-84. doi: 10.1016/j.molmed.2015.03.002. Epub 2015 Apr 4. Trends Mol Med. 2015. PMID: 25851173 Free PMC article. Review.
-
Genomic insights into mite phylogeny, fitness, development, and reproduction.BMC Genomics. 2019 Dec 9;20(1):954. doi: 10.1186/s12864-019-6281-1. BMC Genomics. 2019. PMID: 31818245 Free PMC article.
-
Research Progress on Mono-ADP-Ribosyltransferases in Human Cell Biology.Front Cell Dev Biol. 2022 May 16;10:864101. doi: 10.3389/fcell.2022.864101. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35652091 Free PMC article. Review.
References
-
- Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. - PubMed
-
- Fernandez-Costa JM, Llamusi MB, Garcia-Lopez A, Artero R. Alternative splicing regulation by Muscleblind proteins: from development to disease. Biological reviews of the Cambridge Philosophical Society. 2011;86:947–958. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

