Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton
- PMID: 25371507
- PMCID: PMC4321542
- DOI: 10.1093/jxb/eru437
Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton
Abstract
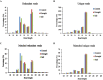
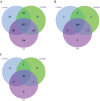
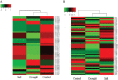
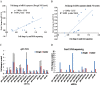
Drought and salinity are two major environmental factors adversely affecting plant growth and productivity. However, the regulatory mechanism is unknown. In this study, the potential roles of small regulatory microRNAs (miRNAs) in cotton response to those stresses were investigated. Using next-generation deep sequencing, a total of 337 miRNAs with precursors were identified, comprising 289 known miRNAs and 48 novel miRNAs. Of these miRNAs, 155 miRNAs were expressed differentially. Target prediction, Gene Ontology (GO)-based functional classification, and Kyoto Encyclopedia of Genes and Genomes (KEGG)-based functional enrichment show that these miRNAs might play roles in response to salinity and drought stresses through targeting a series of stress-related genes. Degradome sequencing analysis showed that at least 55 predicted target genes were further validated to be regulated by 60 miRNAs. CitationRank-based literature mining was employed to determinhe the importance of genes related to drought and salinity stress. The NAC, MYB, and MAPK families were ranked top under the context of drought and salinity, indicating their important roles for the plant to combat drought and salinity stress. According to target prediction, a series of cotton miRNAs are associated with these top-ranked genes, including miR164, miR172, miR396, miR1520, miR6158, ghr-n24, ghr-n56, and ghr-n59. Interestingly, 163 cotton miRNAs were also identified to target 210 genes that are important in fibre development. These results will contribute to cotton stress-resistant breeding as well as understanding fibre development.
Keywords: Cotton; deep sequencing; drought; fibre; literature mining; microRNA; salinity..
© The Author 2014. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

Similar articles
-
High-throughput deep sequencing shows that microRNAs play important roles in switchgrass responses to drought and salinity stress.Plant Biotechnol J. 2014 Apr;12(3):354-66. doi: 10.1111/pbi.12142. Epub 2013 Nov 28. Plant Biotechnol J. 2014. PMID: 24283289
-
High throughput deep degradome sequencing reveals microRNAs and their targets in response to drought stress in mulberry (Morus alba).PLoS One. 2017 Feb 24;12(2):e0172883. doi: 10.1371/journal.pone.0172883. eCollection 2017. PLoS One. 2017. PMID: 28235056 Free PMC article.
-
Small RNA sequencing identifies miRNA roles in ovule and fibre development.Plant Biotechnol J. 2015 Apr;13(3):355-69. doi: 10.1111/pbi.12296. Epub 2015 Jan 9. Plant Biotechnol J. 2015. PMID: 25572837
-
MicroRNAs in cotton: an open world needs more exploration.Planta. 2015 Jun;241(6):1303-12. doi: 10.1007/s00425-015-2282-8. Epub 2015 Apr 5. Planta. 2015. PMID: 25841643 Review.
-
MicroRNA: a new target for improving plant tolerance to abiotic stress.J Exp Bot. 2015 Apr;66(7):1749-61. doi: 10.1093/jxb/erv013. Epub 2015 Feb 19. J Exp Bot. 2015. PMID: 25697792 Free PMC article. Review.
Cited by
-
MicroRNA398: A Master Regulator of Plant Development and Stress Responses.Int J Mol Sci. 2022 Sep 16;23(18):10803. doi: 10.3390/ijms231810803. Int J Mol Sci. 2022. PMID: 36142715 Free PMC article. Review.
-
Regulatory network established by transcription factors transmits drought stress signals in plant.Stress Biol. 2022 Jul 14;2(1):26. doi: 10.1007/s44154-022-00048-z. Stress Biol. 2022. PMID: 37676542 Free PMC article. Review.
-
Non-coding RNAs and transposable elements in plant genomes: emergence, regulatory mechanisms and roles in plant development and stress responses.Planta. 2019 Jul;250(1):23-40. doi: 10.1007/s00425-019-03166-7. Epub 2019 Apr 16. Planta. 2019. PMID: 30993403 Review.
-
The Accumulation of miRNAs Differentially Modulated by Drought Stress Is Affected by Grafting in Grapevine.Plant Physiol. 2017 Apr;173(4):2180-2195. doi: 10.1104/pp.16.01119. Epub 2017 Feb 24. Plant Physiol. 2017. PMID: 28235889 Free PMC article.
-
ccNET: Database of co-expression networks with functional modules for diploid and polyploid Gossypium.Nucleic Acids Res. 2017 Jan 4;45(D1):D1090-D1099. doi: 10.1093/nar/gkw910. Epub 2016 Oct 7. Nucleic Acids Res. 2017. PMID: 28053168 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

