Synaptic, transcriptional and chromatin genes disrupted in autism
- PMID: 25363760
- PMCID: PMC4402723
- DOI: 10.1038/nature13772
Synaptic, transcriptional and chromatin genes disrupted in autism
Abstract
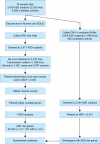
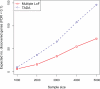
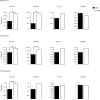
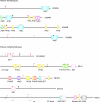
The genetic architecture of autism spectrum disorder involves the interplay of common and rare variants and their impact on hundreds of genes. Using exome sequencing, here we show that analysis of rare coding variation in 3,871 autism cases and 9,937 ancestry-matched or parental controls implicates 22 autosomal genes at a false discovery rate (FDR) < 0.05, plus a set of 107 autosomal genes strongly enriched for those likely to affect risk (FDR < 0.30). These 107 genes, which show unusual evolutionary constraint against mutations, incur de novo loss-of-function mutations in over 5% of autistic subjects. Many of the genes implicated encode proteins for synaptic formation, transcriptional regulation and chromatin-remodelling pathways. These include voltage-gated ion channels regulating the propagation of action potentials, pacemaking and excitability-transcription coupling, as well as histone-modifying enzymes and chromatin remodellers-most prominently those that mediate post-translational lysine methylation/demethylation modifications of histones.
Figures




Similar articles
-
The contribution of de novo coding mutations to autism spectrum disorder.Nature. 2014 Nov 13;515(7526):216-21. doi: 10.1038/nature13908. Epub 2014 Oct 29. Nature. 2014. PMID: 25363768 Free PMC article.
-
Recurrent Mutations of Chromatin-Remodeling Genes and Kinase Receptors in Pheochromocytomas and Paragangliomas.Clin Cancer Res. 2016 May 1;22(9):2301-10. doi: 10.1158/1078-0432.CCR-15-1841. Epub 2015 Dec 23. Clin Cancer Res. 2016. PMID: 26700204 Free PMC article.
-
Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma.Nature. 2012 Jan 29;482(7384):226-31. doi: 10.1038/nature10833. Nature. 2012. PMID: 22286061
-
Recent progresses in molecular genetics of autism spectrum disorders.Yi Chuan. 2015 Sep;37(9):845-54. doi: 10.16288/j.yczz.15-281. Yi Chuan. 2015. PMID: 26399524 Review.
-
From the genetic architecture to synaptic plasticity in autism spectrum disorder.Nat Rev Neurosci. 2015 Sep;16(9):551-63. doi: 10.1038/nrn3992. Nat Rev Neurosci. 2015. PMID: 26289574 Review.
Cited by
-
Cancer-driving mutations are enriched in genic regions intolerant to germline variation.Sci Adv. 2022 Aug 26;8(34):eabo6371. doi: 10.1126/sciadv.abo6371. Epub 2022 Aug 26. Sci Adv. 2022. PMID: 36026442 Free PMC article.
-
α2δ-3 Is Required for Rapid Transsynaptic Homeostatic Signaling.Cell Rep. 2016 Sep 13;16(11):2875-2888. doi: 10.1016/j.celrep.2016.08.030. Cell Rep. 2016. PMID: 27626659 Free PMC article.
-
KnockoffTrio: A knockoff framework for the identification of putative causal variants in genome-wide association studies with trio design.Am J Hum Genet. 2022 Oct 6;109(10):1761-1776. doi: 10.1016/j.ajhg.2022.08.013. Epub 2022 Sep 22. Am J Hum Genet. 2022. PMID: 36150388 Free PMC article.
-
Autism-like Deficits in Shank3-Deficient Mice Are Rescued by Targeting Actin Regulators.Cell Rep. 2015 Jun 9;11(9):1400-1413. doi: 10.1016/j.celrep.2015.04.064. Epub 2015 May 28. Cell Rep. 2015. PMID: 26027926 Free PMC article.
-
BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription.Am J Hum Genet. 2016 Aug 4;99(2):253-74. doi: 10.1016/j.ajhg.2016.05.030. Epub 2016 Jul 21. Am J Hum Genet. 2016. PMID: 27453576 Free PMC article.
References
-
- Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2011;156B:255–274. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- MH077139/MH/NIMH NIH HHS/United States
- U01MH100233/MH/NIMH NIH HHS/United States
- R01 MH077139/MH/NIMH NIH HHS/United States
- MH095034/MH/NIMH NIH HHS/United States
- MR/L010305/1/MRC_/Medical Research Council/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- P30 HD015052/HD/NICHD NIH HHS/United States
- R01 NS073601/NS/NINDS NIH HHS/United States
- WT091310/WT_/Wellcome Trust/United Kingdom
- U01MH100239/MH/NIMH NIH HHS/United States
- R01MH083565/MH/NIMH NIH HHS/United States
- P50 HD055751/HD/NICHD NIH HHS/United States
- R01MH089208/MH/NIMH NIH HHS/United States
- R01 MH094400/MH/NIMH NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- U01 MH100233/MH/NIMH NIH HHS/United States
- WT098051/WT_/Wellcome Trust/United Kingdom
- R01 MH083565/MH/NIMH NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- RC2MH089952/MH/NIMH NIH HHS/United States
- R01 MH089208/MH/NIMH NIH HHS/United States
- R01 MH095034/MH/NIMH NIH HHS/United States
- UL1TR000445/TR/NCATS NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- R01 MH097849/MH/NIMH NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- U01 MH100229/MH/NIMH NIH HHS/United States
- 091986/Wellcome Trust/United Kingdom
- G0500870/MRC_/Medical Research Council/United Kingdom
- R01 MH061009/MH/NIMH NIH HHS/United States
- RC2 MH089952/MH/NIMH NIH HHS/United States
- U01 MH100209/MH/NIMH NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- U01 MH100239/MH/NIMH NIH HHS/United States
- U01MH100209/MH/NIMH NIH HHS/United States
- 5UL1 RR024975/RR/NCRR NIH HHS/United States
- U01MH100229/MH/NIMH NIH HHS/United States
- R01 MH095797/MH/NIMH NIH HHS/United States
- P30 HD15052/HD/NICHD NIH HHS/United States
- MH089482/MH/NIMH NIH HHS/United States
- R01 MH089482/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

