Structural basis for microRNA targeting
- PMID: 25359968
- PMCID: PMC4313529
- DOI: 10.1126/science.1258040
Structural basis for microRNA targeting
Abstract
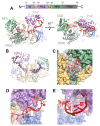
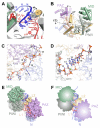
MicroRNAs (miRNAs) control expression of thousands of genes in plants and animals. miRNAs function by guiding Argonaute proteins to complementary sites in messenger RNAs (mRNAs) targeted for repression. We determined crystal structures of human Argonaute-2 (Ago2) bound to a defined guide RNA with and without target RNAs representing miRNA recognition sites. These structures suggest a stepwise mechanism, in which Ago2 primarily exposes guide nucleotides (nt) 2 to 5 for initial target pairing. Pairing to nt 2 to 5 promotes conformational changes that expose nt 2 to 8 and 13 to 16 for further target recognition. Interactions with the guide-target minor groove allow Ago2 to interrogate target RNAs in a sequence-independent manner, whereas an adenosine binding-pocket opposite guide nt 1 further facilitates target recognition. Spurious slicing of miRNA targets is avoided through an inhibitory coordination of one catalytic magnesium ion. These results explain the conserved nucleotide-pairing patterns in animal miRNA target sites first observed over two decades ago.
Copyright © 2014, American Association for the Advancement of Science.
Figures




Comment in
-
RNA. Complete pairing not needed.Science. 2014 Oct 31;346(6209):542-3. doi: 10.1126/science.1262123. Science. 2014. PMID: 25359948 Free PMC article.
Similar articles
-
Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets.Elife. 2015 Sep 11;4:e07646. doi: 10.7554/eLife.07646. Elife. 2015. PMID: 26359634 Free PMC article.
-
Diversity, expression and mRNA targeting abilities of Argonaute-targeting miRNAs among selected vascular plants.BMC Genomics. 2014 Dec 2;15(1):1049. doi: 10.1186/1471-2164-15-1049. BMC Genomics. 2014. PMID: 25443390 Free PMC article.
-
Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2.EMBO J. 2019 Jul 1;38(13):e101153. doi: 10.15252/embj.2018101153. Epub 2019 Apr 26. EMBO J. 2019. PMID: 31268608 Free PMC article.
-
miRNA Targeting: Growing beyond the Seed.Trends Genet. 2019 Mar;35(3):215-222. doi: 10.1016/j.tig.2018.12.005. Epub 2019 Jan 9. Trends Genet. 2019. PMID: 30638669 Free PMC article. Review.
-
AGO unchained: Canonical and non-canonical roles of Argonaute proteins in mammals.Front Biosci (Landmark Ed). 2020 Jan 1;25(1):1-42. doi: 10.2741/4793. Front Biosci (Landmark Ed). 2020. PMID: 31585876 Free PMC article. Review.
Cited by
-
Divergent target recognition by coexpressed 5'-isomiRs of miR-142-3p and selective viral mimicry.RNA. 2015 Sep;21(9):1606-20. doi: 10.1261/rna.048876.114. Epub 2015 Jul 2. RNA. 2015. PMID: 26137849 Free PMC article.
-
Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute.Nat Commun. 2016 Jun 21;7:11846. doi: 10.1038/ncomms11846. Nat Commun. 2016. PMID: 27325485 Free PMC article.
-
miR-30e-5p represses angiogenesis and metastasis by directly targeting AEG-1 in squamous cell carcinoma of the head and neck.Cancer Sci. 2020 Feb;111(2):356-368. doi: 10.1111/cas.14259. Epub 2019 Dec 30. Cancer Sci. 2020. PMID: 31778279 Free PMC article.
-
Neuronal miR-138 Represses HSV-2 Lytic Infection by Regulating Viral and Host Genes with Mechanistic Differences from HSV-1.J Virol. 2022 May 11;96(9):e0034922. doi: 10.1128/jvi.00349-22. Epub 2022 Apr 11. J Virol. 2022. PMID: 35404085 Free PMC article.
-
Structural Basis for Target-Directed MicroRNA Degradation.Mol Cell. 2019 Sep 19;75(6):1243-1255.e7. doi: 10.1016/j.molcel.2019.06.019. Epub 2019 Jul 25. Mol Cell. 2019. PMID: 31353209 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

