Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E
- PMID: 25339359
- PMCID: PMC4256913
- DOI: 10.1182/blood-2014-07-588806
Isolation and transcriptome analyses of human erythroid progenitors: BFU-E and CFU-E
Abstract
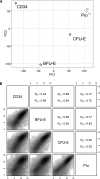
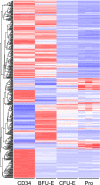
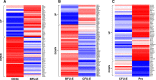
Burst-forming unit-erythroid (BFU-E) and colony-forming unit-erythroid (CFU-E) cells are erythroid progenitors traditionally defined by colony assays. We developed a flow cytometry-based strategy for isolating human BFU-E and CFU-E cells based on the changes in expression of cell surface markers during in vitro erythroid cell culture. BFU-E and CFU-E are characterized by CD45(+)GPA(-)IL-3R(-)CD34(+)CD36(-)CD71(low) and CD45(+)GPA(-)IL-3R(-)CD34(-)CD36(+)CD71(high) phenotypes, respectively. Colony assays validated phenotypic assignment giving rise to BFU-E and CFU-E colonies, both at a purity of ∼90%. The BFU-E colony forming ability of CD45(+)GPA(-)IL-3R(-)CD34(+)CD36(-)CD71(low) cells required stem cell factor and erythropoietin, while the CFU-E colony forming ability of CD45(+)GPA(-)IL-3R(-)CD34(-)CD36(+)CD71(high) cells required only erythropoietin. Bioinformatic analysis of the RNA-sequencing data revealed unique transcriptomes at each differentiation stage. The sorting strategy was validated in uncultured primary cells isolated from bone marrow, cord blood, and peripheral blood, indicating that marker expression is not an artifact of in vitro cell culture, but represents an in vivo characteristic of erythroid progenitor populations. The ability to isolate highly pure human BFU-E and CFU-E progenitors will enable detailed cellular and molecular characterization of these distinct progenitor populations and define their contribution to disordered erythropoiesis in inherited and acquired hematologic disease. Our data provides an important resource for future studies of human erythropoiesis.
© 2014 by The American Society of Hematology.
Figures




Similar articles
-
Flow Cytometry (FCM) Analysis and Fluorescence-Activated Cell Sorting (FACS) of Erythroid Cells.Methods Mol Biol. 2018;1698:153-174. doi: 10.1007/978-1-4939-7428-3_9. Methods Mol Biol. 2018. PMID: 29076089
-
Mast cell growth factor modulates CD36 antigen expression on erythroid progenitors from human bone marrow and peripheral blood associated with ongoing differentiation.Blood. 1994 Jul 1;84(1):59-64. Blood. 1994. PMID: 7517219
-
Comprehensive phenotyping of erythropoiesis in human bone marrow: Evaluation of normal and ineffective erythropoiesis.Am J Hematol. 2021 Sep 1;96(9):1064-1076. doi: 10.1002/ajh.26247. Epub 2021 Jun 3. Am J Hematol. 2021. PMID: 34021930 Free PMC article.
-
Cell surface antigen expression in human erythroid progenitors: erythroid and megakaryocytic markers.Leuk Lymphoma. 1994 May;13(5-6):401-9. doi: 10.3109/10428199409049629. Leuk Lymphoma. 1994. PMID: 8069185 Review.
-
Characterization, regulation, and targeting of erythroid progenitors in normal and disordered human erythropoiesis.Curr Opin Hematol. 2017 May;24(3):159-166. doi: 10.1097/MOH.0000000000000328. Curr Opin Hematol. 2017. PMID: 28099275 Free PMC article. Review.
Cited by
-
β-thalassemias: paradigmatic diseases for scientific discoveries and development of innovative therapies.Haematologica. 2015 Apr;100(4):418-30. doi: 10.3324/haematol.2014.114827. Haematologica. 2015. PMID: 25828088 Free PMC article. Review.
-
Erythroid-intrinsic activation of TLR8 impairs erythropoiesis in inherited anemia.Nat Commun. 2024 Jul 6;15(1):5678. doi: 10.1038/s41467-024-50066-w. Nat Commun. 2024. PMID: 38971858 Free PMC article.
-
Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa, Asia and Oceania.Nat Commun. 2019 Dec 16;10(1):5732. doi: 10.1038/s41467-019-13480-z. Nat Commun. 2019. PMID: 31844061 Free PMC article.
-
Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis.Cell. 2018 Mar 22;173(1):90-103.e19. doi: 10.1016/j.cell.2018.02.036. Epub 2018 Mar 15. Cell. 2018. PMID: 29551269 Free PMC article.
-
The role of TGFβ in hematopoiesis and myeloid disorders.Leukemia. 2019 May;33(5):1076-1089. doi: 10.1038/s41375-019-0420-1. Epub 2019 Feb 28. Leukemia. 2019. PMID: 30816330 Free PMC article. Review.
References
-
- McLeod DL, Shreeve MM, Axelrad AA. Improved plasma culture system for production of erythrocytic colonies in vitro: quantitative assay method for CFU-E. Blood. 1974;44(4):517–534. - PubMed
-
- Iscove NN, Sieber F, Winterhalter KH. Erythroid colony formation in cultures of mouse and human bone marrow: analysis of the requirement for erythropoietin by gel filtration and affinity chromatography on agarose-concanavalin A. J Cell Physiol. 1974;83(2):309–320. - PubMed
-
- Moriyama Y, Fisher JW. Effects of testosterone and erythropoietin on erythroid colony formation in human bone marrow cultures. Blood. 1975;45(5):665–670. - PubMed
-
- Gregory CJ, Eaves AC. Human marrow cells capable of erythropoietic differentiation in vitro: definition of three erythroid colony responses. Blood. 1977;49(6):855–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

