Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile
- PMID: 25337874
- PMCID: PMC4354891
- DOI: 10.1038/nature13828
Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile
Abstract
The gastrointestinal tracts of mammals are colonized by hundreds of microbial species that contribute to health, including colonization resistance against intestinal pathogens. Many antibiotics destroy intestinal microbial communities and increase susceptibility to intestinal pathogens. Among these, Clostridium difficile, a major cause of antibiotic-induced diarrhoea, greatly increases morbidity and mortality in hospitalized patients. Which intestinal bacteria provide resistance to C. difficile infection and their in vivo inhibitory mechanisms remain unclear. Here we correlate loss of specific bacterial taxa with development of infection, by treating mice with different antibiotics that result in distinct microbiota changes and lead to varied susceptibility to C. difficile. Mathematical modelling augmented by analyses of the microbiota of hospitalized patients identifies resistance-associated bacteria common to mice and humans. Using these platforms, we determine that Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium, is associated with resistance to C. difficile infection and, upon administration, enhances resistance to infection in a secondary bile acid dependent fashion. Using a workflow involving mouse models, clinical studies, metagenomic analyses, and mathematical modelling, we identify a probiotic candidate that corrects a clinically relevant microbiome deficiency. These findings have implications for the rational design of targeted antimicrobials as well as microbiome-based diagnostics and therapeutics for individuals at risk of C. difficile infection.
Figures






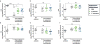

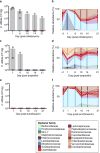

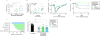

Comment in
-
Infection: microbiota reconstitution for resistance to Clostridium difficile infection--fight fire with fire?Nat Rev Gastroenterol Hepatol. 2015 Jan;12(1):4. doi: 10.1038/nrgastro.2014.194. Epub 2014 Nov 11. Nat Rev Gastroenterol Hepatol. 2015. PMID: 25385229 No abstract available.
-
Microbial bile acid metabolic clusters: the bouncers at the bar.Cell Host Microbe. 2014 Nov 12;16(5):551-2. doi: 10.1016/j.chom.2014.10.015. Epub 2014 Nov 12. Cell Host Microbe. 2014. PMID: 25525784 Free PMC article.
-
Dysfunctional families: Clostridium scindens and secondary bile acids inhibit the growth of Clostridium difficile.Cell Metab. 2015 Jan 6;21(1):9-10. doi: 10.1016/j.cmet.2014.12.016. Cell Metab. 2015. PMID: 25565200 Free PMC article.
-
Systematic discovery of probiotics.Nat Biotechnol. 2015 Jan;33(1):47-9. doi: 10.1038/nbt.3111. Nat Biotechnol. 2015. PMID: 25574637 No abstract available.
Similar articles
-
Dysfunctional families: Clostridium scindens and secondary bile acids inhibit the growth of Clostridium difficile.Cell Metab. 2015 Jan 6;21(1):9-10. doi: 10.1016/j.cmet.2014.12.016. Cell Metab. 2015. PMID: 25565200 Free PMC article.
-
Functional Intestinal Bile Acid 7α-Dehydroxylation by Clostridium scindens Associated with Protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model.Front Cell Infect Microbiol. 2016 Dec 20;6:191. doi: 10.3389/fcimb.2016.00191. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 28066726 Free PMC article.
-
Microbial bile acid metabolic clusters: the bouncers at the bar.Cell Host Microbe. 2014 Nov 12;16(5):551-2. doi: 10.1016/j.chom.2014.10.015. Epub 2014 Nov 12. Cell Host Microbe. 2014. PMID: 25525784 Free PMC article.
-
Role of the intestinal microbiota in resistance to colonization by Clostridium difficile.Gastroenterology. 2014 May;146(6):1547-53. doi: 10.1053/j.gastro.2014.01.059. Epub 2014 Feb 4. Gastroenterology. 2014. PMID: 24503131 Free PMC article. Review.
-
Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract.Anaerobe. 2016 Oct;41:44-50. doi: 10.1016/j.anaerobe.2016.05.003. Epub 2016 May 7. Anaerobe. 2016. PMID: 27163871 Free PMC article. Review.
Cited by
-
Protection from Lethal Clostridioides difficile Infection via Intraspecies Competition for Cogerminant.mBio. 2021 Mar 30;12(2):e00522-21. doi: 10.1128/mBio.00522-21. mBio. 2021. PMID: 33785619 Free PMC article.
-
Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens.Science. 2016 Apr 29;352(6285):535-8. doi: 10.1126/science.aad9382. Science. 2016. PMID: 27126035 Free PMC article. Review.
-
Identification of a microbial sub-community from the feral chicken gut that reduces Salmonella colonization and improves gut health in a gnotobiotic chicken model.Microbiol Spectr. 2024 Mar 5;12(3):e0162123. doi: 10.1128/spectrum.01621-23. Epub 2024 Feb 5. Microbiol Spectr. 2024. PMID: 38315031 Free PMC article.
-
Cancer and the microbiota.Science. 2015 Apr 3;348(6230):80-6. doi: 10.1126/science.aaa4972. Science. 2015. PMID: 25838377 Free PMC article. Review.
-
The impact of dietary fibers on Clostridioides difficile infection in a mouse model.Front Cell Infect Microbiol. 2022 Nov 9;12:1028267. doi: 10.3389/fcimb.2022.1028267. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36439215 Free PMC article.
References
-
- Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. - PubMed
-
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. - PubMed
Supplementary references (Methods)
-
- Chen X, Katchar K, Goldsmith JD, Nanthakumar N, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–1992. - PubMed
-
- Wells JE, Williams KB, Whitehead TR, Heuman DM, Hylemon PB. Development and application of a polymerase chain reaction assay for the detection and enumeration of bile acid 7α-dehydroxylating bacteria in human feces. Clinica Chimica Acta. 2003;331:127–134. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- P01 CA023766/CA/NCI NIH HHS/United States
- T32GM07739/GM/NIGMS NIH HHS/United States
- R01 AI42135/AI/NIAID NIH HHS/United States
- DP2 OD008440/OD/NIH HHS/United States
- R37 AI039031/AI/NIAID NIH HHS/United States
- U54 CA148967/CA/NCI NIH HHS/United States
- DP2OD008440/OD/NIH HHS/United States
- K23 AI095398/AI/NIAID NIH HHS/United States
- T32 CA009149/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- AI95706/AI/NIAID NIH HHS/United States
- R01 AI095706/AI/NIAID NIH HHS/United States
- R01 AI042135/AI/NIAID NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

