The life cycle and pathogenesis of human cytomegalovirus infection: lessons from proteomics
- PMID: 25327590
- PMCID: PMC4604749
- DOI: 10.1586/14789450.2014.971116
The life cycle and pathogenesis of human cytomegalovirus infection: lessons from proteomics
Abstract
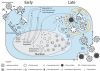
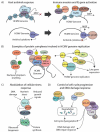
Viruses have coevolved with their hosts, acquiring strategies to subvert host cellular pathways for effective viral replication and spread. Human cytomegalovirus (HCMV), a widely-spread β-herpesvirus, is a major cause of birth defects and opportunistic infections in HIV-1/AIDS patients. HCMV displays an intricate system-wide modulation of the human cell proteome. An impressive array of virus-host protein interactions occurs throughout the infection. To investigate the virus life cycle, proteomics has recently become a significant component of virology studies. Here, we review the mass spectrometry-based proteomics approaches used in HCMV studies, as well as their contribution to understanding the HCMV life cycle and the virus-induced changes to host cells. The importance of the biological insights gained from these studies clearly demonstrate the impact that proteomics has had and can continue to have on understanding HCMV biology and identifying new therapeutic targets.
Keywords: CMV; mass spectrometry; post-translational modifications; protein–protein interactions; proteomics; virus–host interactions.
Figures


Similar articles
-
Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection.Nat Commun. 2020 Feb 10;11(1):806. doi: 10.1038/s41467-020-14586-5. Nat Commun. 2020. PMID: 32041945 Free PMC article.
-
Manipulation of host pathways by human cytomegalovirus: insights from genome-wide studies.Semin Immunopathol. 2014 Nov;36(6):651-8. doi: 10.1007/s00281-014-0443-7. Epub 2014 Sep 27. Semin Immunopathol. 2014. PMID: 25260940 Review.
-
Nitric Oxide Circumvents Virus-Mediated Metabolic Regulation during Human Cytomegalovirus Infection.mBio. 2020 Dec 15;11(6):e02630-20. doi: 10.1128/mBio.02630-20. mBio. 2020. PMID: 33323506 Free PMC article.
-
Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment.J Virol. 2015 Mar;89(6):3049-61. doi: 10.1128/JVI.03416-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552713 Free PMC article.
-
Mechanisms of human cytomegalovirus infection with a focus on epidermal growth factor receptor interactions.Rev Med Virol. 2017 Nov;27(6). doi: 10.1002/rmv.1955. Epub 2017 Oct 11. Rev Med Virol. 2017. PMID: 29024283 Review.
Cited by
-
The host exosome pathway underpins biogenesis of the human cytomegalovirus virion.Elife. 2020 Sep 10;9:e58288. doi: 10.7554/eLife.58288. Elife. 2020. PMID: 32910773 Free PMC article.
-
"Congenital cytomegalovirus in Sub-Saharan Africa-a narrative review with practice recommendations".Front Public Health. 2024 May 15;12:1359663. doi: 10.3389/fpubh.2024.1359663. eCollection 2024. Front Public Health. 2024. PMID: 38813410 Free PMC article. Review.
-
A Portrait of the Human Organelle Proteome In Space and Time during Cytomegalovirus Infection.Cell Syst. 2016 Oct 26;3(4):361-373.e6. doi: 10.1016/j.cels.2016.08.012. Epub 2016 Sep 15. Cell Syst. 2016. PMID: 27641956 Free PMC article.
-
Multiple modes of antigen exposure induce clonotypically diverse epitope-specific CD8+ T cells across multiple tissues in nonhuman primates.PLoS Pathog. 2022 Jul 7;18(7):e1010611. doi: 10.1371/journal.ppat.1010611. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35797339 Free PMC article.
-
Human Cytomegalovirus Manipulates Syntaxin 6 for Successful Trafficking and Subsequent Infection of Monocytes.J Virol. 2022 Jul 27;96(14):e0081922. doi: 10.1128/jvi.00819-22. Epub 2022 Jul 11. J Virol. 2022. PMID: 35862696 Free PMC article.
References
-
- Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Reviews in medical virology. 2010;20:202–213. doi:10.1002/rmv.655. - PubMed
-
- Sinzger C, Digel M, Jahn G. Cytomegalovirus cell tropism. Current topics in microbiology and immunology. 2008;325:63–83. - PubMed
-
- Reeves M, Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Current topics in microbiology and immunology. 2008;325:297–313. - PubMed
-
- Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Reviews in medical virology. 2007;17:253–276. doi:10.1002/rmv.535. - PubMed
-
- Lanzieri TM, Dollard SC, Bialek SR, Grosse SD. Systematic review of the birth prevalence of congenital cytomegalovirus infection in developing countries. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 2014;22:44–48. doi:10.1016/j.ijid.2013.12.010. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
