The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling
- PMID: 25317274
- PMCID: PMC4169610
- DOI: 10.3402/jev.v3.24858
The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling
Abstract
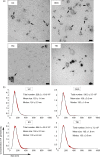
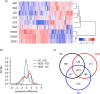
Despite an enormous interest in the role of extracellular vesicles, including exosomes, in cancer and their use as biomarkers for diagnosis, prognosis, drug response and recurrence, there is no consensus on dependable isolation protocols. We provide a comparative evaluation of 4 exosome isolation protocols for their usability, yield and purity, and their impact on downstream omics approaches for biomarker discovery. OptiPrep density gradient centrifugation outperforms ultracentrifugation and ExoQuick and Total Exosome Isolation precipitation in terms of purity, as illustrated by the highest number of CD63-positive nanovesicles, the highest enrichment in exosomal marker proteins and a lack of contaminating proteins such as extracellular Argonaute-2 complexes. The purest exosome fractions reveal a unique mRNA profile enriched for translation, ribosome, mitochondrion and nuclear lumen function. Our results demonstrate that implementation of high purification techniques is a prerequisite to obtain reliable omics data and identify exosome-specific functions and biomarkers.
Keywords: ExoQuick; OptiPrep; exosomes; extracellular vesicles; omics; ultracentrifugation.
Figures






Similar articles
-
Exosome Isolation by Ultracentrifugation and Precipitation and Techniques for Downstream Analyses.Curr Protoc Cell Biol. 2020 Sep;88(1):e110. doi: 10.1002/cpcb.110. Curr Protoc Cell Biol. 2020. PMID: 32633898 Free PMC article.
-
Comparison of serum exosome isolation methods for microRNA profiling.Clin Biochem. 2014 Jan;47(1-2):135-8. doi: 10.1016/j.clinbiochem.2013.10.020. Epub 2013 Oct 29. Clin Biochem. 2014. PMID: 24183884
-
Exosome-like vesicles in uterine aspirates: a comparison of ultracentrifugation-based isolation protocols.J Transl Med. 2016 Jun 18;14(1):180. doi: 10.1186/s12967-016-0935-4. J Transl Med. 2016. PMID: 27317346 Free PMC article.
-
Proteomic analysis of exosomal cargo: the challenge of high purity vesicle isolation.Mol Biosyst. 2016 Apr 26;12(5):1407-19. doi: 10.1039/c6mb00082g. Mol Biosyst. 2016. PMID: 27030573 Review.
-
Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes.Methods. 2015 Oct 1;87:3-10. doi: 10.1016/j.ymeth.2015.02.019. Epub 2015 Mar 10. Methods. 2015. PMID: 25766927 Review.
Cited by
-
Modulation of Immune Responses by Extracellular Vesicles From Retinal Pigment Epithelium.Invest Ophthalmol Vis Sci. 2016 Aug 1;57(10):4101-7. doi: 10.1167/iovs.15-18353. Invest Ophthalmol Vis Sci. 2016. PMID: 27537259 Free PMC article.
-
From cerebral ischemia towards myocardial, renal, and hepatic ischemia: Exosomal miRNAs as a general concept of intercellular communication in ischemia-reperfusion injury.Mol Ther Nucleic Acids. 2022 Aug 24;29:900-922. doi: 10.1016/j.omtn.2022.08.032. eCollection 2022 Sep 13. Mol Ther Nucleic Acids. 2022. PMID: 36159596 Free PMC article. Review.
-
Exosomes as the source of biomarkers of metabolic diseases.Ann Pediatr Endocrinol Metab. 2016 Sep;21(3):119-125. doi: 10.6065/apem.2016.21.3.119. Epub 2016 Sep 30. Ann Pediatr Endocrinol Metab. 2016. PMID: 27777903 Free PMC article. Review.
-
Protocol for serum exosomal miRNAs analysis in prostate cancer patients treated with radiotherapy.J Transl Med. 2018 Aug 13;16(1):223. doi: 10.1186/s12967-018-1592-6. J Transl Med. 2018. PMID: 30103771 Free PMC article.
-
Cortactin and fascin-1 regulate extracellular vesicle release by controlling endosomal trafficking or invadopodia formation and function.Sci Rep. 2018 Oct 23;8(1):15606. doi: 10.1038/s41598-018-33868-z. Sci Rep. 2018. PMID: 30353022 Free PMC article.
References
-
- Simons M, Raposo G. Exosomes – vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81. - PubMed
-
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. - PubMed
-
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 Chapter 3:Unit 3. - PubMed
-
- Cantin R, Diou J, Belanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods. 2008;338:21–30. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

