Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity
- PMID: 25311841
- PMCID: PMC4259884
- DOI: 10.1007/s13238-014-0104-6
Coronavirus membrane-associated papain-like proteases induce autophagy through interacting with Beclin1 to negatively regulate antiviral innate immunity
Abstract
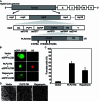
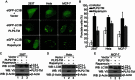
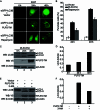
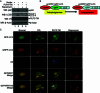
Autophagy plays important roles in modulating viral replication and antiviral immune response. Coronavirus infection is associated with the autophagic process, however, little is known about the mechanisms of autophagy induction and its contribution to coronavirus regulation of host innate responses. Here, we show that the membrane-associated papain-like protease PLP2 (PLP2-TM) of coronaviruses acts as a novel autophagy-inducing protein. Intriguingly, PLP2-TM induces incomplete autophagy process by increasing the accumulation of autophagosomes but blocking the fusion of autophagosomes with lysosomes. Furthermore, PLP2-TM interacts with the key autophagy regulators, LC3 and Beclin1, and promotes Beclin1 interaction with STING, the key regulator for antiviral IFN signaling. Finally, knockdown of Beclin1 partially reverses PLP2-TM's inhibitory effect on innate immunity which resulting in decreased coronavirus replication. These results suggested that coronavirus papain-like protease induces incomplete autophagy by interacting with Beclin1, which in turn modulates coronavirus replication and antiviral innate immunity.
Figures



Similar articles
-
Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling.PLoS One. 2012;7(2):e30802. doi: 10.1371/journal.pone.0030802. Epub 2012 Feb 1. PLoS One. 2012. PMID: 22312431 Free PMC article.
-
Deubiquitinase function of arterivirus papain-like protease 2 suppresses the innate immune response in infected host cells.Proc Natl Acad Sci U S A. 2013 Feb 26;110(9):E838-47. doi: 10.1073/pnas.1218464110. Epub 2013 Feb 11. Proc Natl Acad Sci U S A. 2013. PMID: 23401522 Free PMC article.
-
Chikungunya triggers an autophagic process which promotes viral replication.Virol J. 2011 Sep 8;8:432. doi: 10.1186/1743-422X-8-432. Virol J. 2011. PMID: 21902836 Free PMC article.
-
Message in a bottle: lessons learned from antagonism of STING signalling during RNA virus infection.Cytokine Growth Factor Rev. 2014 Dec;25(6):669-79. doi: 10.1016/j.cytogfr.2014.08.004. Epub 2014 Aug 24. Cytokine Growth Factor Rev. 2014. PMID: 25212897 Free PMC article. Review.
-
Strategies employed by viruses to manipulate autophagy.Prog Mol Biol Transl Sci. 2020;172:203-237. doi: 10.1016/bs.pmbts.2020.01.004. Epub 2020 Feb 10. Prog Mol Biol Transl Sci. 2020. PMID: 32620243 Review.
Cited by
-
Activation of STING Signaling Pathway Effectively Blocks Human Coronavirus Infection.J Virol. 2021 May 24;95(12):e00490-21. doi: 10.1128/JVI.00490-21. Print 2021 May 24. J Virol. 2021. PMID: 33789998 Free PMC article.
-
Autophagy: A Key Regulator of Homeostasis and Disease: An Overview of Molecular Mechanisms and Modulators.Cells. 2022 Jul 22;11(15):2262. doi: 10.3390/cells11152262. Cells. 2022. PMID: 35892559 Free PMC article. Review.
-
The ER stress sensor IRE1 and MAP kinase ERK modulate autophagy induction in cells infected with coronavirus infectious bronchitis virus.Virology. 2019 Jul;533:34-44. doi: 10.1016/j.virol.2019.05.002. Epub 2019 May 2. Virology. 2019. PMID: 31082732 Free PMC article.
-
MCPIP1 negatively regulate cellular antiviral innate immune responses through DUB and disruption of TRAF3-TBK1-IKKε complex.Biochem Biophys Res Commun. 2018 Sep 5;503(2):830-836. doi: 10.1016/j.bbrc.2018.06.083. Epub 2018 Jun 27. Biochem Biophys Res Commun. 2018. PMID: 29920243 Free PMC article.
-
Autophagy, Unfolded Protein Response, and Neuropilin-1 Cross-Talk in SARS-CoV-2 Infection: What Can Be Learned from Other Coronaviruses.Int J Mol Sci. 2021 Jun 1;22(11):5992. doi: 10.3390/ijms22115992. Int J Mol Sci. 2021. PMID: 34206057 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

