sc-PDB: a 3D-database of ligandable binding sites--10 years on
- PMID: 25300483
- PMCID: PMC4384012
- DOI: 10.1093/nar/gku928
sc-PDB: a 3D-database of ligandable binding sites--10 years on
Abstract
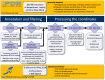
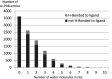
The sc-PDB database (available at http://bioinfo-pharma.u-strasbg.fr/scPDB/) is a comprehensive and up-to-date selection of ligandable binding sites of the Protein Data Bank. Sites are defined from complexes between a protein and a pharmacological ligand. The database provides the all-atom description of the protein, its ligand, their binding site and their binding mode. Currently, the sc-PDB archive registers 9283 binding sites from 3678 unique proteins and 5608 unique ligands. The sc-PDB database was publicly launched in 2004 with the aim of providing structure files suitable for computational approaches to drug design, such as docking. During the last 10 years we have improved and standardized the processes for (i) identifying binding sites, (ii) correcting structures, (iii) annotating protein function and ligand properties and (iv) characterizing their binding mode. This paper presents the latest enhancements in the database, specifically pertaining to the representation of molecular interaction and to the similarity between ligand/protein binding patterns. The new website puts emphasis in pictorial analysis of data.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
sc-PDB: a database for identifying variations and multiplicity of 'druggable' binding sites in proteins.Bioinformatics. 2011 May 1;27(9):1324-6. doi: 10.1093/bioinformatics/btr120. Epub 2011 Mar 12. Bioinformatics. 2011. PMID: 21398668
-
sc-PDB: an annotated database of druggable binding sites from the Protein Data Bank.J Chem Inf Model. 2006 Mar-Apr;46(2):717-27. doi: 10.1021/ci050372x. J Chem Inf Model. 2006. PMID: 16563002
-
sc-PDB-Frag: a database of protein-ligand interaction patterns for Bioisosteric replacements.J Chem Inf Model. 2014 Jul 28;54(7):1908-18. doi: 10.1021/ci500282c. Epub 2014 Jul 17. J Chem Inf Model. 2014. PMID: 24991975
-
The Art of Compiling Protein Binding Site Ensembles.Mol Inform. 2016 Dec;35(11-12):593-598. doi: 10.1002/minf.201600043. Epub 2016 May 30. Mol Inform. 2016. PMID: 27870245 Review.
-
Molecular modeling of hydration in drug design.Curr Opin Drug Discov Devel. 2007 May;10(3):275-80. Curr Opin Drug Discov Devel. 2007. PMID: 17554853 Review.
Cited by
-
Turbocharging protein binding site prediction with geometric attention, inter-resolution transfer learning, and homology-based augmentation.BMC Bioinformatics. 2024 Sep 20;25(1):306. doi: 10.1186/s12859-024-05923-2. BMC Bioinformatics. 2024. PMID: 39304807 Free PMC article.
-
Formulation and in-silico study of meclizine ointment as anti-eczema.In Silico Pharmacol. 2022 Aug 30;10(1):15. doi: 10.1007/s40203-022-00129-x. eCollection 2022. In Silico Pharmacol. 2022. PMID: 36062215 Free PMC article.
-
Estimating the Similarity between Protein Pockets.Int J Mol Sci. 2022 Oct 18;23(20):12462. doi: 10.3390/ijms232012462. Int J Mol Sci. 2022. PMID: 36293316 Free PMC article. Review.
-
Computational methods and tools for binding site recognition between proteins and small molecules: from classical geometrical approaches to modern machine learning strategies.J Comput Aided Mol Des. 2019 Oct;33(10):887-903. doi: 10.1007/s10822-019-00235-7. Epub 2019 Oct 18. J Comput Aided Mol Des. 2019. PMID: 31628659
-
Unexpected similarity between HIV-1 reverse transcriptase and tumor necrosis factor binding sites revealed by computer vision.J Cheminform. 2021 Nov 23;13(1):90. doi: 10.1186/s13321-021-00567-3. J Cheminform. 2021. PMID: 34814950 Free PMC article.
References
-
- Ripphausen P., Nisius B., Peltason L., Bajorath J. Quo Vadis, virtual screening? A comprehensive survey of prospective applications. J. Med. Chem. 2010;53:8461–8467. - PubMed
-
- Hopkins A.L., Groom C.R. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. - PubMed
-
- Edfeldt F.N., Folmer R.H., Breeze A.L. Fragment screening to predict druggability (ligandability) and lead discovery success. Drug Discov. Today. 2011;16:284–287. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

