Clonal dynamics of native haematopoiesis
- PMID: 25296256
- PMCID: PMC4408613
- DOI: 10.1038/nature13824
Clonal dynamics of native haematopoiesis
Abstract
It is currently thought that life-long blood cell production is driven by the action of a small number of multipotent haematopoietic stem cells. Evidence supporting this view has been largely acquired through the use of functional assays involving transplantation. However, whether these mechanisms also govern native non-transplant haematopoiesis is entirely unclear. Here we have established a novel experimental model in mice where cells can be uniquely and genetically labelled in situ to address this question. Using this approach, we have performed longitudinal analyses of clonal dynamics in adult mice that reveal unprecedented features of native haematopoiesis. In contrast to what occurs following transplantation, steady-state blood production is maintained by the successive recruitment of thousands of clones, each with a minimal contribution to mature progeny. Our results demonstrate that a large number of long-lived progenitors, rather than classically defined haematopoietic stem cells, are the main drivers of steady-state haematopoiesis during most of adulthood. Our results also have implications for understanding the cellular origin of haematopoietic disease.
Conflict of interest statement
The authors declare no competing financial interests.
Figures








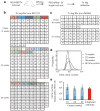

Comment in
-
Sleeping beauty wakes up the clonal succession model for homeostatic hematopoiesis.Cell Stem Cell. 2014 Dec 4;15(6):677-8. doi: 10.1016/j.stem.2014.11.015. Cell Stem Cell. 2014. PMID: 25479744
Similar articles
-
Clonal analysis of lineage fate in native haematopoiesis.Nature. 2018 Jan 11;553(7687):212-216. doi: 10.1038/nature25168. Epub 2018 Jan 3. Nature. 2018. PMID: 29323290 Free PMC article.
-
Clonal tracking of haematopoietic cells: insights and clinical implications.Br J Haematol. 2021 Mar;192(5):819-831. doi: 10.1111/bjh.17175. Epub 2020 Nov 20. Br J Haematol. 2021. PMID: 33216985 Free PMC article. Review.
-
Developmental fate of hematopoietic stem cells: the study of individual hematopoietic clones at the level of antigen-responsive B lymphocytes.Hematol J. 2003;4(2):146-50. doi: 10.1038/sj.thj.6200239. Hematol J. 2003. PMID: 12750734
-
Deciphering cell states and genealogies of human haematopoiesis.Nature. 2024 Mar;627(8003):389-398. doi: 10.1038/s41586-024-07066-z. Epub 2024 Jan 22. Nature. 2024. PMID: 38253266 Free PMC article.
-
Do haematopoietic stem cells age?Nat Rev Immunol. 2020 Mar;20(3):196-202. doi: 10.1038/s41577-019-0236-2. Epub 2019 Nov 18. Nat Rev Immunol. 2020. PMID: 31740804 Free PMC article. Review.
Cited by
-
Hematopoietic Stem Cell Metabolism during Development and Aging.Dev Cell. 2020 Jul 20;54(2):239-255. doi: 10.1016/j.devcel.2020.06.029. Dev Cell. 2020. PMID: 32693057 Free PMC article. Review.
-
Feedback mechanisms control coexistence in a stem cell model of acute myeloid leukaemia.J Theor Biol. 2016 Jul 21;401:43-53. doi: 10.1016/j.jtbi.2016.04.002. Epub 2016 Apr 27. J Theor Biol. 2016. PMID: 27130539 Free PMC article.
-
Awakening the HSC: Dynamic Modeling of HSC Maintenance Unravels Regulation of the TP53 Pathway and Quiescence.Front Physiol. 2020 Jul 31;11:848. doi: 10.3389/fphys.2020.00848. eCollection 2020. Front Physiol. 2020. PMID: 32848827 Free PMC article.
-
HSC Contribution in Making Steady-State Blood.Immunity. 2016 Sep 20;45(3):464-466. doi: 10.1016/j.immuni.2016.09.002. Immunity. 2016. PMID: 27653597 Free PMC article. Review.
-
Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells.J Clin Invest. 2015 May;125(5):2007-20. doi: 10.1172/JCI78124. Epub 2015 Apr 13. J Clin Invest. 2015. PMID: 25866973 Free PMC article.
References
-
- Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. - PubMed
-
- Kondo M, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. - PubMed
-
- Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. - PubMed
-
- Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. - PubMed
-
- Bystrykh LV, Verovskaya E, Zwart E, Broekhuis M, de Haan G. Counting stem cells: methodological constraints. Nature Methods. 2012;9:567–574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

