Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library
- PMID: 25294943
- PMCID: PMC4197710
- DOI: 10.1242/dev.111054
Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library
Abstract
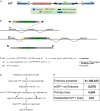
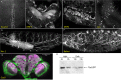
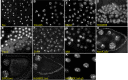
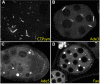
Although we now have a wealth of information on the transcription patterns of all the genes in the Drosophila genome, much less is known about the properties of the encoded proteins. To provide information on the expression patterns and subcellular localisations of many proteins in parallel, we have performed a large-scale protein trap screen using a hybrid piggyBac vector carrying an artificial exon encoding yellow fluorescent protein (YFP) and protein affinity tags. From screening 41 million embryos, we recovered 616 verified independent YFP-positive lines representing protein traps in 374 genes, two-thirds of which had not been tagged in previous P element protein trap screens. Over 20 different research groups then characterized the expression patterns of the tagged proteins in a variety of tissues and at several developmental stages. In parallel, we purified many of the tagged proteins from embryos using the affinity tags and identified co-purifying proteins by mass spectrometry. The fly stocks are publicly available through the Kyoto Drosophila Genetics Resource Center. All our data are available via an open access database (Flannotator), which provides comprehensive information on the expression patterns, subcellular localisations and in vivo interaction partners of the trapped proteins. Our resource substantially increases the number of available protein traps in Drosophila and identifies new markers for cellular organelles and structures.
Keywords: Affinity purification; Cytoophidia; Live imaging; Protein trap; piggyBac.
© 2014. Published by The Company of Biologists Ltd.
Figures


Similar articles
-
Subcellular localisations of the CPTI collection of YFP-tagged proteins in Drosophila embryos.Development. 2014 Oct;141(20):4006-17. doi: 10.1242/dev.111310. Development. 2014. PMID: 25294944 Free PMC article.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article. Review.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
Cited by
-
Protein trap: a new Swiss army knife for geneticists?Mol Biol Rep. 2020 Feb;47(2):1445-1458. doi: 10.1007/s11033-019-05181-z. Epub 2019 Nov 14. Mol Biol Rep. 2020. PMID: 31728729 Review.
-
NanoDam identifies Homeobrain (ARX) and Scarecrow (NKX2.1) as conserved temporal factors in the Drosophila central brain and visual system.Dev Cell. 2022 May 9;57(9):1193-1207.e7. doi: 10.1016/j.devcel.2022.04.008. Epub 2022 Apr 27. Dev Cell. 2022. PMID: 35483359 Free PMC article.
-
miRNA-mediated gene silencing in Drosophila larval development involves GW182-dependent and independent mechanisms.EMBO J. 2024 Dec;43(23):6161-6179. doi: 10.1038/s44318-024-00249-4. Epub 2024 Sep 25. EMBO J. 2024. PMID: 39322759 Free PMC article.
-
RhoGAP19D inhibits Cdc42 laterally to control epithelial cell shape and prevent invasion.J Cell Biol. 2021 Apr 5;220(4):e202009116. doi: 10.1083/jcb.202009116. J Cell Biol. 2021. PMID: 33646271 Free PMC article.
-
SILAC-iPAC: a quantitative method for distinguishing genuine from non-specific components of protein complexes by parallel affinity capture.J Proteomics. 2015 Feb 6;115:143-56. doi: 10.1016/j.jprot.2014.12.006. Epub 2014 Dec 20. J Proteomics. 2015. PMID: 25534881 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BB/F010303/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 103792/WT_/Wellcome Trust/United Kingdom
- 096144/WT_/Wellcome Trust/United Kingdom
- BB/C503903/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 092096/WT_/Wellcome Trust/United Kingdom
- BB/F014287/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/G02247X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H016473/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 049818/WT_/Wellcome Trust/United Kingdom
- 99234/WT_/Wellcome Trust/United Kingdom
- A14492/CRUK_/Cancer Research UK/United Kingdom
- 076739/WT_/Wellcome Trust/United Kingdom
- BB/D009324/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 080007/WT_/Wellcome Trust/United Kingdom
- 091911/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

