CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors
- PMID: 25294932
- PMCID: PMC4210312
- DOI: 10.1073/pnas.1405266111
CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors
Abstract
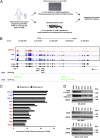
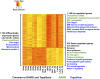
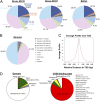
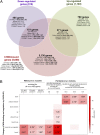
Truncating mutations of chromodomain helicase DNA-binding protein 8 (CHD8), and of many other genes with diverse functions, are strong-effect risk factors for autism spectrum disorder (ASD), suggesting multiple mechanisms of pathogenesis. We explored the transcriptional networks that CHD8 regulates in neural progenitor cells (NPCs) by reducing its expression and then integrating transcriptome sequencing (RNA sequencing) with genome-wide CHD8 binding (ChIP sequencing). Suppressing CHD8 to levels comparable with the loss of a single allele caused altered expression of 1,756 genes, 64.9% of which were up-regulated. CHD8 showed widespread binding to chromatin, with 7,324 replicated sites that marked 5,658 genes. Integration of these data suggests that a limited array of direct regulatory effects of CHD8 produced a much larger network of secondary expression changes. Genes indirectly down-regulated (i.e., without CHD8-binding sites) reflect pathways involved in brain development, including synapse formation, neuron differentiation, cell adhesion, and axon guidance, whereas CHD8-bound genes are strongly associated with chromatin modification and transcriptional regulation. Genes associated with ASD were strongly enriched among indirectly down-regulated loci (P < 10(-8)) and CHD8-bound genes (P = 0.0043), which align with previously identified coexpression modules during fetal development. We also find an intriguing enrichment of cancer-related gene sets among CHD8-bound genes (P < 10(-10)). In vivo suppression of chd8 in zebrafish produced macrocephaly comparable to that of humans with inactivating mutations. These data indicate that heterozygous disruption of CHD8 precipitates a network of gene-expression changes involved in neurodevelopmental pathways in which many ASD-associated genes may converge on shared mechanisms of pathogenesis.
Keywords: CHD8; ChIP-seq; NPCs; RNA-seq; autism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells.Mol Autism. 2017 Mar 20;8:11. doi: 10.1186/s13229-017-0124-1. eCollection 2017. Mol Autism. 2017. PMID: 28321286 Free PMC article.
-
The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment.Nat Commun. 2015 Mar 10;6:6404. doi: 10.1038/ncomms7404. Nat Commun. 2015. PMID: 25752243 Free PMC article.
-
The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes.Transl Psychiatry. 2015 May 19;5(5):e568. doi: 10.1038/tp.2015.62. Transl Psychiatry. 2015. PMID: 25989142 Free PMC article.
-
Mutations and Modeling of the Chromatin Remodeler CHD8 Define an Emerging Autism Etiology.Front Neurosci. 2015 Dec 17;9:477. doi: 10.3389/fnins.2015.00477. eCollection 2015. Front Neurosci. 2015. PMID: 26733790 Free PMC article. Review.
-
The Mechanisms of CHD8 in Neurodevelopment and Autism Spectrum Disorders.Genes (Basel). 2021 Jul 26;12(8):1133. doi: 10.3390/genes12081133. Genes (Basel). 2021. PMID: 34440307 Free PMC article. Review.
Cited by
-
Epigenetic changes in the developing brain: Effects on behavior.Proc Natl Acad Sci U S A. 2015 Jun 2;112(22):6789-95. doi: 10.1073/pnas.1501482112. Proc Natl Acad Sci U S A. 2015. PMID: 26034282 Free PMC article. No abstract available.
-
Genome-wide variant analysis of simplex autism families with an integrative clinical-bioinformatics pipeline.Cold Spring Harb Mol Case Stud. 2015 Oct;1(1):a000422. doi: 10.1101/mcs.a000422. Cold Spring Harb Mol Case Stud. 2015. PMID: 27148569 Free PMC article.
-
Review: Cancer and neurodevelopmental disorders: multi-scale reasoning and computational guide.Front Cell Dev Biol. 2024 Jul 2;12:1376639. doi: 10.3389/fcell.2024.1376639. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39015651 Free PMC article. Review.
-
Reply to: "Correspondence to bipolar disorder-iPSC derived neural progenitor cells exhibit dysregulation of store-operated Ca2+ entry and accelerated differentiation" by Yde Ohki and colleagues.Mol Psychiatry. 2024 Jul 31. doi: 10.1038/s41380-024-02673-8. Online ahead of print. Mol Psychiatry. 2024. PMID: 39085393 No abstract available.
-
Association of genetic variants with autism spectrum disorder in Japanese children revealed by targeted sequencing.Front Genet. 2024 Aug 30;15:1352480. doi: 10.3389/fgene.2024.1352480. eCollection 2024. Front Genet. 2024. PMID: 39280100 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

