Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26
- PMID: 25211075
- PMCID: PMC7104937
- DOI: 10.1016/j.chom.2014.08.009
Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26
Abstract
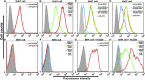
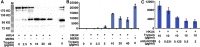
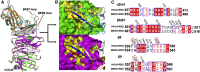
The recently reported Middle East respiratory syndrome coronavirus (MERS-CoV) is phylogenetically closely related to the bat coronaviruses (BatCoVs) HKU4 and HKU5. However, the evolutionary pathway of MERS-CoV is still unclear. A receptor binding domain (RBD) in the MERS-CoV envelope-embedded spike protein specifically engages human CD26 (hCD26) to initiate viral entry. The high sequence identity in the viral spike protein prompted us to investigate if HKU4 and HKU5 can recognize hCD26 for cell entry. We found that HKU4-RBD, but not HKU5-RBD, binds to hCD26, and pseudotyped viruses embedding HKU4 spike can infect cells via hCD26 recognition. The structure of the HKU4-RBD/hCD26 complex revealed a hCD26-binding mode similar overall to that observed for MERS-RBD. HKU4-RBD, however, is less adapted to hCD26 than MERS-RBD, explaining its lower affinity for receptor binding. Our findings support a bat origin for MERS-CoV and indicate the need for surveillance of HKU4-related viruses in bats.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



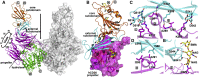
Similar articles
-
Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein.Virology. 2017 Jul;507:101-109. doi: 10.1016/j.virol.2017.04.016. Epub 2017 Apr 19. Virology. 2017. PMID: 28432925 Free PMC article.
-
Detection and full genome characterization of two beta CoV viruses related to Middle East respiratory syndrome from bats in Italy.Virol J. 2017 Dec 19;14(1):239. doi: 10.1186/s12985-017-0907-1. Virol J. 2017. PMID: 29258555 Free PMC article.
-
Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus.Proc Natl Acad Sci U S A. 2014 Aug 26;111(34):12516-21. doi: 10.1073/pnas.1405889111. Epub 2014 Aug 11. Proc Natl Acad Sci U S A. 2014. PMID: 25114257 Free PMC article.
-
Unraveling the Epidemiology, Geographical Distribution, and Genomic Evolution of Potentially Lethal Coronaviruses (SARS, MERS, and SARS CoV-2).Front Cell Infect Microbiol. 2020 Aug 27;10:499. doi: 10.3389/fcimb.2020.00499. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32974224 Free PMC article. Review.
-
The lethal internal face of the coronaviruses: Kidney tropism of the SARS, MERS, and COVID19 viruses.IUBMB Life. 2021 Aug;73(8):1005-1015. doi: 10.1002/iub.2516. Epub 2021 Jun 30. IUBMB Life. 2021. PMID: 34118117 Free PMC article. Review.
Cited by
-
Structure, Function, and Evolution of Coronavirus Spike Proteins.Annu Rev Virol. 2016 Sep 29;3(1):237-261. doi: 10.1146/annurev-virology-110615-042301. Epub 2016 Aug 25. Annu Rev Virol. 2016. PMID: 27578435 Free PMC article. Review.
-
Full Genome Nobecovirus Sequences From Malagasy Fruit Bats Define a Unique Evolutionary History for This Coronavirus Clade.Front Public Health. 2022 Feb 11;10:786060. doi: 10.3389/fpubh.2022.786060. eCollection 2022. Front Public Health. 2022. PMID: 35223729 Free PMC article.
-
Precise location of two novel linear epitopes on the receptor-binding domain surface of MERS-CoV spike protein recognized by two different monoclonal antibodies.Int J Biol Macromol. 2022 Jan 15;195:609-619. doi: 10.1016/j.ijbiomac.2021.11.192. Epub 2021 Dec 4. Int J Biol Macromol. 2022. PMID: 34871658 Free PMC article.
-
What Have We Learned About Middle East Respiratory Syndrome Coronavirus Emergence in Humans? A Systematic Literature Review.Vector Borne Zoonotic Dis. 2019 Mar;19(3):174-192. doi: 10.1089/vbz.2017.2191. Epub 2019 Jan 24. Vector Borne Zoonotic Dis. 2019. PMID: 30676269 Free PMC article.
-
Global Epidemiology of Bat Coronaviruses.Viruses. 2019 Feb 20;11(2):174. doi: 10.3390/v11020174. Viruses. 2019. PMID: 30791586 Free PMC article. Review.
References
-
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17:20290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

