Functional implications and ubiquitin-dependent degradation of the peptide transporter Ptr2 in Saccharomyces cerevisiae
- PMID: 25172766
- PMCID: PMC4248705
- DOI: 10.1128/EC.00094-14
Functional implications and ubiquitin-dependent degradation of the peptide transporter Ptr2 in Saccharomyces cerevisiae
Abstract
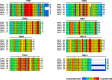
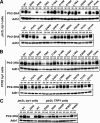
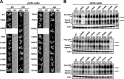
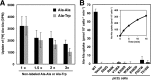
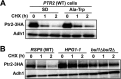
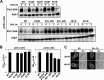
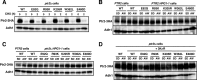
The peptide transporter Ptr2 plays a central role in di- or tripeptide import in Saccharomyces cerevisiae. Although PTR2 transcription has been extensively analyzed in terms of upregulation by the Ubr1-Cup9 circuit, the structural and functional information for this transporter is limited. Here we identified 14 amino acid residues required for peptide import through Ptr2 based on the crystallographic information of Streptococcus thermophilus peptide transporter PepTst and based on the conservation of primary sequences among the proton-dependent oligopeptide transporters (POTs). Expression of Ptr2 carrying one of the 14 mutations of which the corresponding residues of PepTst are involved in peptide recognition, salt bridge interaction, or peptide translocation failed to enable ptr2Δtrp1 cell growth in alanyl-tryptophan (Ala-Trp) medium. We observed that Ptr2 underwent rapid degradation after cycloheximide treatment (half-life, approximately 1 h), and this degradation depended on Rsp5 ubiquitin ligase. The ubiquitination of Ptr2 most likely occurs at the N-terminal lysines 16, 27, and 34. Simultaneous substitution of arginine for the three lysines fully prevented Ptr2 degradation. Ptr2 mutants of the presumed peptide-binding site (E92Q, R93K, K205R, W362L, and E480D) exhibited severe defects in peptide import and were subjected to Rsp5-dependent degradation when cells were moved to Ala-Trp medium, whereas, similar to what occurs in the wild-type Ptr2, mutant proteins of the intracellular gate were upregulated. These results suggest that Ptr2 undergoes quality control and the defects in peptide binding and the concomitant conformational change render Ptr2 subject to efficient ubiquitination and subsequent degradation.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Amino acids induce peptide uptake via accelerated degradation of CUP9, the transcriptional repressor of the PTR2 peptide transporter.J Biol Chem. 2008 Oct 24;283(43):28958-68. doi: 10.1074/jbc.M803980200. Epub 2008 Aug 15. J Biol Chem. 2008. PMID: 18708352 Free PMC article.
-
Pressure-induced endocytic degradation of the Saccharomyces cerevisiae low-affinity tryptophan permease Tat1 is mediated by Rsp5 ubiquitin ligase and functionally redundant PPxY motif proteins.Eukaryot Cell. 2013 Jul;12(7):990-7. doi: 10.1128/EC.00049-13. Epub 2013 May 10. Eukaryot Cell. 2013. PMID: 23666621 Free PMC article.
-
GGA2- and ubiquitin-dependent trafficking of Arn1, the ferrichrome transporter of Saccharomyces cerevisiae.Mol Biol Cell. 2007 May;18(5):1790-802. doi: 10.1091/mbc.e06-09-0861. Epub 2007 Mar 7. Mol Biol Cell. 2007. PMID: 17344478 Free PMC article.
-
The ubiquitin code of yeast permease trafficking.Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
-
Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae.Mol Membr Biol. 2001 Jan-Mar;18(1):105-12. Mol Membr Biol. 2001. PMID: 11396605 Review.
Cited by
-
Study of the Plasma Membrane Proteome Dynamics Reveals Novel Targets of the Nitrogen Regulation in Yeast.Mol Cell Proteomics. 2017 Sep;16(9):1652-1668. doi: 10.1074/mcp.M116.064923. Epub 2017 Jul 5. Mol Cell Proteomics. 2017. PMID: 28679684 Free PMC article.
-
AMPK-Mediated Regulation of Alpha-Arrestins and Protein Trafficking.Int J Mol Sci. 2019 Jan 25;20(3):515. doi: 10.3390/ijms20030515. Int J Mol Sci. 2019. PMID: 30691068 Free PMC article. Review.
-
Yeast α-arrestin Art2 is the key regulator of ubiquitylation-dependent endocytosis of plasma membrane vitamin B1 transporters.PLoS Biol. 2019 Oct 28;17(10):e3000512. doi: 10.1371/journal.pbio.3000512. eCollection 2019 Oct. PLoS Biol. 2019. PMID: 31658248 Free PMC article.
-
Titanium dioxide nanoparticles impart protection from ultraviolet irradiation to fermenting yeast cells.Biochem Biophys Rep. 2022 Feb 4;30:101221. doi: 10.1016/j.bbrep.2022.101221. eCollection 2022 Jul. Biochem Biophys Rep. 2022. PMID: 35685033 Free PMC article.
-
α-Arrestins and Their Functions: From Yeast to Human Health.Int J Mol Sci. 2022 Apr 30;23(9):4988. doi: 10.3390/ijms23094988. Int J Mol Sci. 2022. PMID: 35563378 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

