Ubiquitin-binding site 2 of ataxin-3 prevents its proteasomal degradation by interacting with Rad23
- PMID: 25144244
- PMCID: PMC4237202
- DOI: 10.1038/ncomms5638
Ubiquitin-binding site 2 of ataxin-3 prevents its proteasomal degradation by interacting with Rad23
Abstract
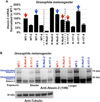
Polyglutamine repeat expansion in ataxin-3 causes neurodegeneration in the most common dominant ataxia, spinocerebellar ataxia type 3 (SCA3). Since reducing levels of disease proteins improves pathology in animals, we investigated how ataxin-3 is degraded. Here we show that, unlike most proteins, ataxin-3 turnover does not require its ubiquitination, but is regulated by ubiquitin-binding site 2 (UbS2) on its N terminus. Mutating UbS2 decreases ataxin-3 protein levels in cultured mammalian cells and in Drosophila melanogaster by increasing its proteasomal turnover. Ataxin-3 interacts with the proteasome-associated proteins Rad23A/B through UbS2. Knockdown of Rad23 in cultured cells and in Drosophila results in lower levels of ataxin-3 protein. Importantly, reducing Rad23 suppresses ataxin-3-dependent degeneration in flies. We present a mechanism for ubiquitination-independent degradation that is impeded by protein interactions with proteasome-associated factors. We conclude that UbS2 is a potential target through which to enhance ataxin-3 degradation for SCA3 therapy.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




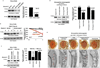

Similar articles
-
Interaction of the polyglutamine protein ataxin-3 with Rad23 regulates toxicity in Drosophila models of Spinocerebellar Ataxia Type 3.Hum Mol Genet. 2017 Apr 15;26(8):1419-1431. doi: 10.1093/hmg/ddx039. Hum Mol Genet. 2017. PMID: 28158474 Free PMC article.
-
A fine balance between Prpf19 and Exoc7 in achieving degradation of aggregated protein and suppression of cell death in spinocerebellar ataxia type 3.Cell Death Dis. 2021 Feb 2;12(2):136. doi: 10.1038/s41419-021-03444-x. Cell Death Dis. 2021. PMID: 33542212 Free PMC article.
-
The deubiquitinase ataxin-3 requires Rad23 and DnaJ-1 for its neuroprotective role in Drosophila melanogaster.Neurobiol Dis. 2015 Oct;82:12-21. doi: 10.1016/j.nbd.2015.05.010. Epub 2015 May 22. Neurobiol Dis. 2015. PMID: 26007638 Free PMC article.
-
Toward therapeutic targets for SCA3: Insight into the role of Machado-Joseph disease protein ataxin-3 in misfolded proteins clearance.Prog Neurobiol. 2015 Sep;132:34-58. doi: 10.1016/j.pneurobio.2015.06.004. Epub 2015 Jun 27. Prog Neurobiol. 2015. PMID: 26123252 Review.
-
The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation.DNA Repair (Amst). 2009 Apr 5;8(4):449-60. doi: 10.1016/j.dnarep.2009.01.005. Epub 2009 Feb 14. DNA Repair (Amst). 2009. PMID: 19223247 Review.
Cited by
-
Reduction of RAD23A extends lifespan and mitigates pathology in TDP-43 mice.bioRxiv [Preprint]. 2024 Sep 14:2024.09.10.612226. doi: 10.1101/2024.09.10.612226. bioRxiv. 2024. PMID: 39314471 Free PMC article. Preprint.
-
ATXN3: a multifunctional protein involved in the polyglutamine disease spinocerebellar ataxia type 3.Expert Rev Mol Med. 2024 Sep 25;26:e19. doi: 10.1017/erm.2024.10. Expert Rev Mol Med. 2024. PMID: 39320846 Free PMC article. Review.
-
A survey of protein interactions and posttranslational modifications that influence the polyglutamine diseases.Front Mol Neurosci. 2022 Sep 14;15:974167. doi: 10.3389/fnmol.2022.974167. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36187346 Free PMC article. Review.
-
Phenotypic defects from the expression of wild-type and pathogenic TATA-binding proteins in new Drosophila models of Spinocerebellar Ataxia Type 17.G3 (Bethesda). 2023 Sep 30;13(10):jkad180. doi: 10.1093/g3journal/jkad180. G3 (Bethesda). 2023. PMID: 37551423 Free PMC article.
-
Androgen receptor polyglutamine expansion drives age-dependent quality control defects and muscle dysfunction.J Clin Invest. 2018 Aug 1;128(8):3630-3641. doi: 10.1172/JCI99042. Epub 2018 Jul 23. J Clin Invest. 2018. PMID: 29809168 Free PMC article.
References
-
- Todi SV, Williams A, Paulson HL. Polyglutamine Repeat Disorders, including Huntington’s Disease. In: Waxman SG, editor. Molecular Neurology. 1st edition. Academic Press; 2007.
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Ann. Rev. Neurosci. 2007;30:575–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

