Synaptic dysregulation in a human iPS cell model of mental disorders
- PMID: 25132547
- PMCID: PMC4501856
- DOI: 10.1038/nature13716
Synaptic dysregulation in a human iPS cell model of mental disorders
Abstract
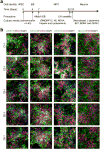
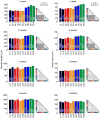
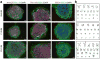
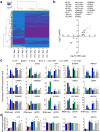
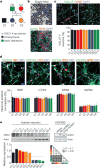
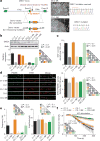
Dysregulated neurodevelopment with altered structural and functional connectivity is believed to underlie many neuropsychiatric disorders, and 'a disease of synapses' is the major hypothesis for the biological basis of schizophrenia. Although this hypothesis has gained indirect support from human post-mortem brain analyses and genetic studies, little is known about the pathophysiology of synapses in patient neurons and how susceptibility genes for mental disorders could lead to synaptic deficits in humans. Genetics of most psychiatric disorders are extremely complex due to multiple susceptibility variants with low penetrance and variable phenotypes. Rare, multiply affected, large families in which a single genetic locus is probably responsible for conferring susceptibility have proven invaluable for the study of complex disorders. Here we generated induced pluripotent stem (iPS) cells from four members of a family in which a frameshift mutation of disrupted in schizophrenia 1 (DISC1) co-segregated with major psychiatric disorders and we further produced different isogenic iPS cell lines via gene editing. We showed that mutant DISC1 causes synaptic vesicle release deficits in iPS-cell-derived forebrain neurons. Mutant DISC1 depletes wild-type DISC1 protein and, furthermore, dysregulates expression of many genes related to synapses and psychiatric disorders in human forebrain neurons. Our study reveals that a psychiatric disorder relevant mutation causes synapse deficits and transcriptional dysregulation in human neurons and our findings provide new insight into the molecular and synaptic etiopathology of psychiatric disorders.
Conflict of interest statement
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Figures



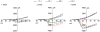


Similar articles
-
Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.Mol Psychiatry. 2021 Apr;26(4):1346-1360. doi: 10.1038/s41380-019-0485-2. Epub 2019 Aug 23. Mol Psychiatry. 2021. PMID: 31444471 Free PMC article.
-
Kinase network dysregulation in a human induced pluripotent stem cell model of DISC1 schizophrenia.Mol Omics. 2019 Jun 10;15(3):173-188. doi: 10.1039/c8mo00173a. Mol Omics. 2019. PMID: 31106784 Free PMC article.
-
DISC1, astrocytes and neuronal maturation: a possible mechanistic link with implications for mental disorders.J Neurochem. 2016 Aug;138(4):518-24. doi: 10.1111/jnc.13663. J Neurochem. 2016. PMID: 27187935
-
Role of DISC1 in Neuronal Trafficking and its Implication in Neuropsychiatric Manifestation and Neurotherapeutics.Neurotherapeutics. 2017 Jul;14(3):623-629. doi: 10.1007/s13311-017-0556-5. Neurotherapeutics. 2017. PMID: 28664299 Free PMC article. Review.
-
Linking neurodevelopmental and synaptic theories of mental illness through DISC1.Nat Rev Neurosci. 2011 Nov 18;12(12):707-22. doi: 10.1038/nrn3120. Nat Rev Neurosci. 2011. PMID: 22095064 Free PMC article. Review.
Cited by
-
The proteome of schizophrenia.NPJ Schizophr. 2015 Mar 4;1:14003. doi: 10.1038/npjschz.2014.3. eCollection 2015. NPJ Schizophr. 2015. PMID: 27336025 Free PMC article. Review.
-
Probing disorders of the nervous system using reprogramming approaches.EMBO J. 2015 Jun 3;34(11):1456-77. doi: 10.15252/embj.201591267. Epub 2015 Apr 29. EMBO J. 2015. PMID: 25925386 Free PMC article. Review.
-
KEOPS complex expression in the frontal cortex in major depression and schizophrenia.World J Biol Psychiatry. 2021 Jul;22(6):446-455. doi: 10.1080/15622975.2020.1821917. Epub 2020 Sep 29. World J Biol Psychiatry. 2021. PMID: 32914678 Free PMC article.
-
Current advancements of modelling schizophrenia using patient-derived induced pluripotent stem cells.Acta Neuropathol Commun. 2022 Dec 16;10(1):183. doi: 10.1186/s40478-022-01460-2. Acta Neuropathol Commun. 2022. PMID: 36527106 Free PMC article. Review.
-
Comparative characterization of human induced pluripotent stem cells (hiPSC) derived from patients with schizophrenia and autism.Transl Psychiatry. 2019 Jul 29;9(1):179. doi: 10.1038/s41398-019-0517-3. Transl Psychiatry. 2019. PMID: 31358727 Free PMC article.
References
-
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. - PubMed
-
- Mirnics K, Middleton FA, Lewis DA, Levitt P. Analysis of complex brain disorders with gene expression microarrays: schizophrenia as a disease of the synapse. Trends Neurosci. 2001;24:479–486. - PubMed
-
- Johnson RD, Oliver PL, Davies KE. SNARE proteins and schizophrenia: linking synaptic and neurodevelopmental hypotheses. Acta Biochim Pol. 2008;55:619–628. - PubMed
-
- Honer WG, Young CE. Presynaptic proteins and schizophrenia. Int Rev Neurobiol. 2004;59:175–199. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R21 MH092740/MH/NIMH NIH HHS/United States
- F31 MH102978/MH/NIMH NIH HHS/United States
- R21 MH087874/MH/NIMH NIH HHS/United States
- R01 AG024984/AG/NIA NIH HHS/United States
- R33 MH087874/MH/NIMH NIH HHS/United States
- NS048271/NS/NINDS NIH HHS/United States
- R37 NS047344/NS/NINDS NIH HHS/United States
- R01 AG045656/AG/NIA NIH HHS/United States
- R56 NS047344/NS/NINDS NIH HHS/United States
- R01 NS048271/NS/NINDS NIH HHS/United States
- R01 NS047344/NS/NINDS NIH HHS/United States
- NS047344/NS/NINDS NIH HHS/United States
- AG045656/AG/NIA NIH HHS/United States
- R01 MH083911/MH/NIMH NIH HHS/United States
- MH102978/MH/NIMH NIH HHS/United States
- R21 ES021957/ES/NIEHS NIH HHS/United States
- MH087874/MH/NIMH NIH HHS/United States
- T32 GM008752/GM/NIGMS NIH HHS/United States
- R01 MH105128/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

