Quantification of crypt and stem cell evolution in the normal and neoplastic human colon
- PMID: 25127143
- PMCID: PMC4471679
- DOI: 10.1016/j.celrep.2014.07.019
Quantification of crypt and stem cell evolution in the normal and neoplastic human colon
Erratum in
-
Quantification of Crypt and Stem Cell Evolution in the Normal and Neoplastic Human Colon.Cell Rep. 2019 May 21;27(8):2524. doi: 10.1016/j.celrep.2019.05.035. Cell Rep. 2019. PMID: 31116993 Free PMC article. No abstract available.
Abstract
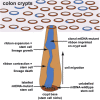
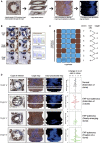
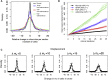
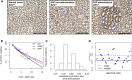
Human intestinal stem cell and crypt dynamics remain poorly characterized because transgenic lineage-tracing methods are impractical in humans. Here, we have circumvented this problem by quantitatively using somatic mtDNA mutations to trace clonal lineages. By analyzing clonal imprints on the walls of colonic crypts, we show that human intestinal stem cells conform to one-dimensional neutral drift dynamics with a "functional" stem cell number of five to six in both normal patients and individuals with familial adenomatous polyposis (germline APC(-/+)). Furthermore, we show that, in adenomatous crypts (APC(-/-)), there is a proportionate increase in both functional stem cell number and the loss/replacement rate. Finally, by analyzing fields of mtDNA mutant crypts, we show that a normal colon crypt divides around once every 30-40 years, and the division rate is increased in adenomas by at least an order of magnitude. These data provide in vivo quantification of human intestinal stem cell and crypt dynamics.
Copyright © 2014 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Comment in
-
Stem cell competition: how speeding mutants beat the rest.EMBO J. 2014 Oct 16;33(20):2277-8. doi: 10.15252/embj.201489823. Epub 2014 Sep 1. EMBO J. 2014. PMID: 25180234 Free PMC article.
Similar articles
-
Enhanced stem cell survival in familial adenomatous polyposis.Am J Pathol. 2004 Apr;164(4):1369-77. doi: 10.1016/S0002-9440(10)63223-3. Am J Pathol. 2004. PMID: 15039224 Free PMC article.
-
Colonic crypt changes during adenoma development in familial adenomatous polyposis: immunohistochemical evidence for expansion of the crypt base cell population.Am J Pathol. 2004 Nov;165(5):1489-98. doi: 10.1016/S0002-9440(10)63407-4. Am J Pathol. 2004. PMID: 15509520 Free PMC article.
-
APC in the regulation of intestinal crypt fission.J Pathol. 1998 Jul;185(3):246-55. doi: 10.1002/(SICI)1096-9896(199807)185:3<246::AID-PATH90>3.0.CO;2-8. J Pathol. 1998. PMID: 9771477
-
Expansion of a mutated clone: from stem cell to tumour.J Clin Pathol. 2008 Feb;61(2):164-71. doi: 10.1136/jcp.2006.044610. Epub 2007 Apr 27. J Clin Pathol. 2008. PMID: 17468295 Review.
-
Measuring stem cell dynamics in the human colon--where there's a wiggle, there's a way.J Pathol. 2014 Nov;234(3):292-5. doi: 10.1002/path.4422. J Pathol. 2014. PMID: 25112223 Free PMC article. Review.
Cited by
-
Characterization of LGR5 stem cells in colorectal adenomas and carcinomas.Sci Rep. 2015 Mar 2;5:8654. doi: 10.1038/srep08654. Sci Rep. 2015. PMID: 25728748 Free PMC article.
-
Implications of Epigenetic Drift in Colorectal Neoplasia.Cancer Res. 2019 Feb 1;79(3):495-504. doi: 10.1158/0008-5472.CAN-18-1682. Epub 2018 Oct 5. Cancer Res. 2019. PMID: 30291105 Free PMC article.
-
Molecular and cellular basis of the dose-rate-dependent adverse effects of radiation exposure in animal models. Part I: Mammary gland and digestive tract.J Radiat Res. 2023 Mar 23;64(2):210-227. doi: 10.1093/jrr/rrad002. J Radiat Res. 2023. PMID: 36773323 Free PMC article. Review.
-
Mature enteroendocrine cells contribute to basal and pathological stem cell dynamics in the small intestine.Am J Physiol Gastrointest Liver Physiol. 2018 Oct 1;315(4):G495-G510. doi: 10.1152/ajpgi.00036.2018. Epub 2018 May 31. Am J Physiol Gastrointest Liver Physiol. 2018. PMID: 29848020 Free PMC article.
-
Multipotent Basal Stem Cells, Maintained in Localized Proximal Niches, Support Directed Long-Ranging Epithelial Flows in Human Prostates.Cell Rep. 2017 Aug 15;20(7):1609-1622. doi: 10.1016/j.celrep.2017.07.061. Cell Rep. 2017. PMID: 28813673 Free PMC article.
References
-
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Cheng H., Bjerknes M., Amar J., Gardiner G. Crypt production in normal and diseased human colonic epithelium. Anat. Rec. 1986;216:44–48. - PubMed
-
- Cheng H., McCulloch C., Bjerknes M. Effects of 30% intestinal resection on whole population cell kinetics of mouse intestinal epithelium. Anat. Rec. 1986;215:35–41. - PubMed
-
- Fellous T.G., McDonald S.A.C., Burkert J., Humphries A., Islam S., De-Alwis N.M.W., Gutierrez-Gonzalez L., Tadrous P.J., Elia G., Kocher H.M. A methodological approach to tracing cell lineage in human epithelial tissues. Stem Cells. 2009;27:1410–1420. - PubMed
-
- Fletcher A.G., Breward C.J., Jonathan Chapman S. Mathematical modeling of monoclonal conversion in the colonic crypt. J. Theor. Biol. 2012;300:118–133. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

