Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense
- PMID: 25126784
- PMCID: PMC4134512
- DOI: 10.1016/j.cell.2014.06.023
Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense
Abstract
DEAD-box helicases play essential roles in RNA metabolism across species, but emerging data suggest that they have additional functions in immunity. Through RNAi screening, we identify an evolutionarily conserved and interferon-independent role for the DEAD-box helicase DDX17 in restricting Rift Valley fever virus (RVFV), a mosquito-transmitted virus in the bunyavirus family that causes severe morbidity and mortality in humans and livestock. Loss of Drosophila DDX17 (Rm62) in cells and flies enhanced RVFV infection. Similarly, depletion of DDX17 but not the related helicase DDX5 increased RVFV replication in human cells. Using crosslinking immunoprecipitation high-throughput sequencing (CLIP-seq), we show that DDX17 binds the stem loops of host pri-miRNA to facilitate their processing and also an essential stem loop in bunyaviral RNA to restrict infection. Thus, DDX17 has dual roles in the recognition of stem loops: in the nucleus for endogenous microRNA (miRNA) biogenesis and in the cytoplasm for surveillance against structured non-self-elements.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
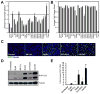
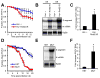
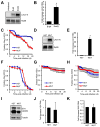
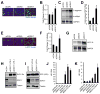
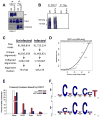
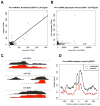
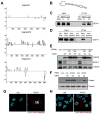
Comment in
-
DDX17: structured RNA recognition drives diverse outputs.Cell Cycle. 2014;13(22):3467-8. doi: 10.4161/15384101.2014.980695. Cell Cycle. 2014. PMID: 25493410 Free PMC article. No abstract available.
Similar articles
-
RNA Helicase DDX17 Inhibits Hepatitis B Virus Replication by Blocking Viral Pregenomic RNA Encapsidation.J Virol. 2021 Sep 9;95(19):e0044421. doi: 10.1128/JVI.00444-21. Epub 2021 Sep 9. J Virol. 2021. PMID: 34287051 Free PMC article.
-
DDX17: structured RNA recognition drives diverse outputs.Cell Cycle. 2014;13(22):3467-8. doi: 10.4161/15384101.2014.980695. Cell Cycle. 2014. PMID: 25493410 Free PMC article. No abstract available.
-
Human DDX17 Unwinds Rift Valley Fever Virus Non-Coding RNAs.Int J Mol Sci. 2020 Dec 23;22(1):54. doi: 10.3390/ijms22010054. Int J Mol Sci. 2020. PMID: 33374561 Free PMC article.
-
The DDX5/Dbp2 subfamily of DEAD-box RNA helicases.Wiley Interdiscip Rev RNA. 2019 Mar;10(2):e1519. doi: 10.1002/wrna.1519. Epub 2018 Dec 2. Wiley Interdiscip Rev RNA. 2019. PMID: 30506978 Free PMC article. Review.
-
DEAD-Box Helicases: Sensors, Regulators, and Effectors for Antiviral Defense.Viruses. 2020 Feb 5;12(2):181. doi: 10.3390/v12020181. Viruses. 2020. PMID: 32033386 Free PMC article. Review.
Cited by
-
Host Cell Restriction Factors of Bunyaviruses and Viral Countermeasures.Viruses. 2021 Apr 28;13(5):784. doi: 10.3390/v13050784. Viruses. 2021. PMID: 33925004 Free PMC article. Review.
-
Defense Mechanisms against Viral Infection in Drosophila: RNAi and Non-RNAi.Viruses. 2018 May 1;10(5):230. doi: 10.3390/v10050230. Viruses. 2018. PMID: 29723993 Free PMC article. Review.
-
RNA Helicase DDX17 Inhibits Hepatitis B Virus Replication by Blocking Viral Pregenomic RNA Encapsidation.J Virol. 2021 Sep 9;95(19):e0044421. doi: 10.1128/JVI.00444-21. Epub 2021 Sep 9. J Virol. 2021. PMID: 34287051 Free PMC article.
-
The RNA helicase DDX5 promotes viral infection via regulating N6-methyladenosine levels on the DHX58 and NFκB transcripts to dampen antiviral innate immunity.PLoS Pathog. 2021 Apr 28;17(4):e1009530. doi: 10.1371/journal.ppat.1009530. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33909701 Free PMC article.
-
The DEAD box RNA helicase DDX42 is an intrinsic inhibitor of positive-strand RNA viruses.EMBO Rep. 2022 Nov 7;23(11):e54061. doi: 10.15252/embr.202154061. Epub 2022 Sep 26. EMBO Rep. 2022. PMID: 36161446 Free PMC article.
References
-
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01AI095500/AI/NIAID NIH HHS/United States
- R56 AI074951/AI/NIAID NIH HHS/United States
- R01 AI095500/AI/NIAID NIH HHS/United States
- U54 AI057168/AI/NIAID NIH HHS/United States
- R01 AI074951/AI/NIAID NIH HHS/United States
- T32 AI007324/AI/NIAID NIH HHS/United States
- R01AI074951/AI/NIAID NIH HHS/United States
- U54AI057168/AI/NIAID NIH HHS/United States
- R01 GM067719/GM/NIGMS NIH HHS/United States
- R01 GM103383/GM/NIGMS NIH HHS/United States
- R01GM103383/GM/NIGMS NIH HHS/United States
- T32AI007324/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

