Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2
- PMID: 25122779
- PMCID: PMC4248901
- DOI: 10.1128/JVI.00970-14
Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2
Abstract
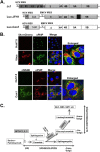
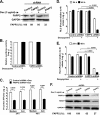
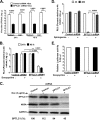
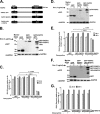
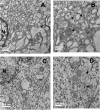
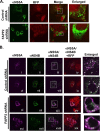
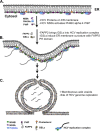
Hepatitis C virus (HCV) assembles its replication complex on cytosolic membrane vesicles often clustered in a membranous web (MW). During infection, HCV NS5A protein activates PI4KIIIα enzyme, causing massive production and redistribution of phosphatidylinositol 4-phosphate (PI4P) lipid to the replication complex. However, the role of PI4P in the HCV life cycle is not well understood. We postulated that PI4P recruits host effectors to modulate HCV genome replication or virus particle production. To test this hypothesis, we generated cell lines for doxycycline-inducible expression of short hairpin RNAs (shRNAs) targeting the PI4P effector, four-phosphate adaptor protein 2 (FAPP2). FAPP2 depletion attenuated HCV infectivity and impeded HCV RNA synthesis. Indeed, FAPP2 has two functional lipid-binding domains specific for PI4P and glycosphingolipids. While expression of the PI4P-binding mutant protein was expected to inhibit HCV replication, a marked drop in replication efficiency was observed unexpectedly with the glycosphingolipid-binding mutant protein. These data suggest that both domains are crucial for the role of FAPP2 in HCV genome replication. We also found that HCV significantly increases the level of some glycosphingolipids, whereas adding these lipids to FAPP2-depleted cells partially rescued replication, further arguing for the importance of glycosphingolipids in HCV RNA synthesis. Interestingly, FAPP2 is redistributed to the replication complex (RC) characterized by HCV NS5A, NS4B, or double-stranded RNA (dsRNA) foci. Additionally, FAPP2 depletion disrupts the RC and alters the colocalization of HCV replicase proteins. Altogether, our study implies that HCV coopts FAPP2 for virus genome replication via PI4P binding and glycosphingolipid transport to the HCV RC.
Importance: Like most viruses with a positive-sense RNA genome, HCV replicates its RNA on remodeled host membranes composed of lipids hijacked from various internal membrane compartments. During infection, HCV induces massive production and retargeting of the PI4P lipid to its replication complex. However, the role of PI4P in HCV replication is not well understood. In this study, we have shown that FAPP2, a PI4P effector and glycosphingolipid-binding protein, is recruited to the HCV replication complex and is required for HCV genome replication and replication complex formation. More importantly, this study demonstrates, for the first time, the crucial role of glycosphingolipids in the HCV life cycle and suggests a link between PI4P and glycosphingolipids in HCV genome replication.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Hepatitis C Virus Subverts Human Choline Kinase-α To Bridge Phosphatidylinositol-4-Kinase IIIα (PI4KIIIα) and NS5A and Upregulates PI4KIIIα Activation, Thereby Promoting the Translocation of the Ternary Complex to the Endoplasmic Reticulum for Viral Replication.J Virol. 2017 Jul 27;91(16):e00355-17. doi: 10.1128/JVI.00355-17. Print 2017 Aug 15. J Virol. 2017. PMID: 28566381 Free PMC article.
-
Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity.PLoS Pathog. 2012;8(3):e1002576. doi: 10.1371/journal.ppat.1002576. Epub 2012 Mar 8. PLoS Pathog. 2012. PMID: 22412376 Free PMC article.
-
Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication.J Virol. 2011 Sep;85(17):8870-83. doi: 10.1128/JVI.00059-11. Epub 2011 Jun 22. J Virol. 2011. PMID: 21697487 Free PMC article.
-
Phosphoinositides in the hepatitis C virus life cycle.Viruses. 2012 Oct 19;4(10):2340-58. doi: 10.3390/v4102340. Viruses. 2012. PMID: 23202467 Free PMC article. Review.
-
Connecting vesicular transport with lipid synthesis: FAPP2.Biochim Biophys Acta. 2012 Aug;1821(8):1089-95. doi: 10.1016/j.bbalip.2012.01.003. Epub 2012 Jan 14. Biochim Biophys Acta. 2012. PMID: 22266015 Free PMC article. Review.
Cited by
-
Glucosylceramide synthase inhibitors prevent replication of SARS-CoV-2 and influenza virus.J Biol Chem. 2021 Jan-Jun;296:100470. doi: 10.1016/j.jbc.2021.100470. Epub 2021 Feb 25. J Biol Chem. 2021. PMID: 33639165 Free PMC article.
-
Entangled in a membranous web: ER and lipid droplet reorganization during hepatitis C virus infection.Curr Opin Cell Biol. 2016 Aug;41:117-24. doi: 10.1016/j.ceb.2016.05.003. Epub 2016 May 27. Curr Opin Cell Biol. 2016. PMID: 27240021 Free PMC article. Review.
-
The emerging roles of retromer and sorting nexins in the life cycle of viruses.Virol Sin. 2022 Jun;37(3):321-330. doi: 10.1016/j.virs.2022.04.014. Epub 2022 May 2. Virol Sin. 2022. PMID: 35513271 Free PMC article. Review.
-
Hepatitis C virus alters the morphology and function of peroxisomes.Front Microbiol. 2023 Sep 21;14:1254728. doi: 10.3389/fmicb.2023.1254728. eCollection 2023. Front Microbiol. 2023. PMID: 37808318 Free PMC article.
-
Hepatitis C Virus Subverts Human Choline Kinase-α To Bridge Phosphatidylinositol-4-Kinase IIIα (PI4KIIIα) and NS5A and Upregulates PI4KIIIα Activation, Thereby Promoting the Translocation of the Ternary Complex to the Endoplasmic Reticulum for Viral Replication.J Virol. 2017 Jul 27;91(16):e00355-17. doi: 10.1128/JVI.00355-17. Print 2017 Aug 15. J Virol. 2017. PMID: 28566381 Free PMC article.
References
-
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. 10.1002/hep.20819. - DOI - PubMed
-
- Bartenschlager R, Lohmann V. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81(Part 7):1631–1648 http://vir.sgmjournals.org/content/81/7/1631.long. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

