The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism
- PMID: 25122757
- PMCID: PMC4183825
- DOI: 10.1074/jbc.M114.590505
The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism
Abstract
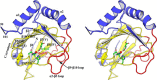
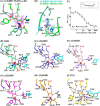
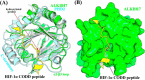
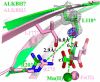
ALKBH7 is the mitochondrial AlkB family member that is required for alkylation- and oxidation-induced programmed necrosis. In contrast to the protective role of other AlkB family members after suffering alkylation-induced DNA damage, ALKBH7 triggers the collapse of mitochondrial membrane potential and promotes cell death. Moreover, genetic ablation of mouse Alkbh7 dramatically increases body weight and fat mass. Here, we present crystal structures of human ALKBH7 in complex with Mn(II) and α-ketoglutarate at 1.35 Å or N-oxalylglycine at 2.0 Å resolution. ALKBH7 possesses the conserved double-stranded β-helix fold that coordinates a catalytically active iron by a conserved HX(D/E) … Xn … H motif. Self-hydroxylation of Leu-110 was observed, indicating that ALKBH7 has the potential to catalyze hydroxylation of its substrate. Unlike other AlkB family members whose substrates are DNA or RNA, ALKBH7 is devoid of the "nucleotide recognition lid" which is essential for binding nucleobases, and thus exhibits a solvent-exposed active site; two loops between β-strands β6 and β7 and between β9 and β10 create a special outer wall of the minor β-sheet of the double-stranded β-helix and form a negatively charged groove. These distinct features suggest that ALKBH7 may act on protein substrate rather than nucleic acids. Taken together, our findings provide a structural basis for understanding the distinct function of ALKBH7 in the AlkB family and offer a foundation for drug design in treating cell death-related diseases and metabolic diseases.
Keywords: ALKB; ALKBH7; Crystal Structure; Dioxygenase; Fatty Acid Metabolism; Necrosis (Necrotic Death); Obesity; Protein Hydroxylase; Self-Hydroxylation; X-ray Crystallography.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




Similar articles
-
ALKBH7 mediates necrosis via rewiring of glyoxal metabolism.Elife. 2020 Aug 14;9:e58573. doi: 10.7554/eLife.58573. Elife. 2020. PMID: 32795389 Free PMC article.
-
Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis.Genes Dev. 2013 May 15;27(10):1089-100. doi: 10.1101/gad.215533.113. Epub 2013 May 10. Genes Dev. 2013. PMID: 23666923 Free PMC article.
-
Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition.J Biol Chem. 2014 Apr 25;289(17):11571-11583. doi: 10.1074/jbc.M113.546168. Epub 2014 Mar 10. J Biol Chem. 2014. PMID: 24616105 Free PMC article.
-
The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond.J Biol Chem. 2015 Aug 21;290(34):20734-20742. doi: 10.1074/jbc.R115.656462. Epub 2015 Jul 7. J Biol Chem. 2015. PMID: 26152727 Free PMC article. Review.
-
The AlkB Family of Fe (II)/Alpha-Ketoglutarate-Dependent Dioxygenases Modulates Embryogenesis through Epigenetic Regulation.Curr Stem Cell Res Ther. 2018;13(2):136-143. doi: 10.2174/1574888X12666171027105532. Curr Stem Cell Res Ther. 2018. PMID: 29076432 Review.
Cited by
-
Multi-substrate selectivity based on key loops and non-homologous domains: new insight into ALKBH family.Cell Mol Life Sci. 2021 Jan;78(1):129-141. doi: 10.1007/s00018-020-03594-9. Epub 2020 Jul 8. Cell Mol Life Sci. 2021. PMID: 32642789 Free PMC article. Review.
-
Spectroscopic and in vitro Investigations of Fe2+ /α-Ketoglutarate-Dependent Enzymes Involved in Nucleic Acid Repair and Modification.Chembiochem. 2022 Jun 3;23(11):e202100605. doi: 10.1002/cbic.202100605. Epub 2022 Feb 15. Chembiochem. 2022. PMID: 35040547 Free PMC article. Review.
-
ALKBH7 mediates necrosis via rewiring of glyoxal metabolism.Elife. 2020 Aug 14;9:e58573. doi: 10.7554/eLife.58573. Elife. 2020. PMID: 32795389 Free PMC article.
-
The crystal structure of the thiocyanate-forming protein from Thlaspi arvense, a kelch protein involved in glucosinolate breakdown.Plant Mol Biol. 2015 Sep;89(1-2):67-81. doi: 10.1007/s11103-015-0351-9. Epub 2015 Aug 11. Plant Mol Biol. 2015. PMID: 26260516
-
Evolutionary History of RNA Modifications at N6-Adenosine Originating from the R-M System in Eukaryotes and Prokaryotes.Biology (Basel). 2022 Jan 28;11(2):214. doi: 10.3390/biology11020214. Biology (Basel). 2022. PMID: 35205080 Free PMC article.
References
-
- Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E. S., Baehrecke E. H., Blagosklonny M. V., El-Deiry W. S., Golstein P., Green D. R., Hengartner M., Knight R. A., Kumar S., Lipton S. A., Malorni W., Nuñez G., Peter M. E., Tschopp J., Yuan J., Piacentini M., Zhivotovsky B., Melino G., and Nomenclature Committee on Cell Death 2009 (2009) Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death. Differ. 16, 3–11 - PMC - PubMed
-
- Dunai Z., Bauer P. I., Mihalik R. (2011) Necroptosis: biochemical, physiological and pathological aspects. Pathol. Oncol. Res. 17, 791–800 - PubMed
-
- Cipriani G., Rapizzi E., Vannacci A., Rizzuto R., Moroni F., Chiarugi A. (2005) Nuclear poly(ADP-ribose) polymerase-1 rapidly triggers mitochondrial dysfunction. J. Biol. Chem. 280, 17227–17234 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

