Diagnostic application of PIK3CA mutation analysis in Chinese esophageal cancer patients
- PMID: 25106743
- PMCID: PMC4149237
- DOI: 10.1186/s13000-014-0153-4
Diagnostic application of PIK3CA mutation analysis in Chinese esophageal cancer patients
Abstract
Background: The PIK3CA gene mutation was found to associate with prognosis and might affect molecular targeted therapy in esophageal carcinoma (EC). The aim of this study is to compare different methods for analyzing the PIK3CA gene mutation in EC.
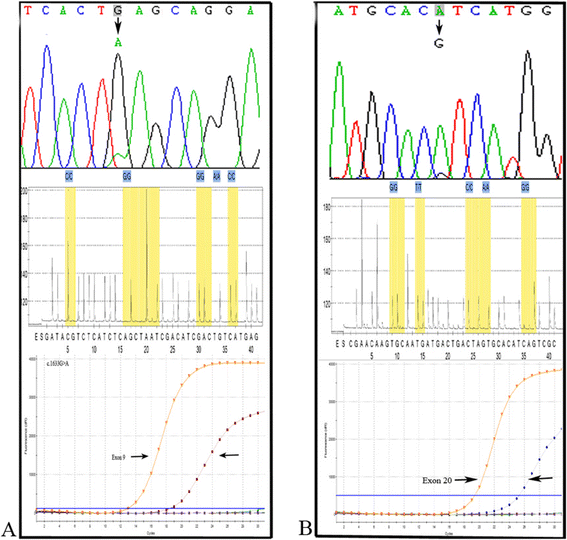
Methods: Genomic DNA was extracted from 106 surgically resected EC patient tissues. The PIK3CA mutation status (exons 9 and 20) were screened by mutant-enrich liquid chip (ME-Liquidchip), Sanger sequencing, and pyrosequencing. And all samples with mutations were independently reassessed using amplification refractory mutation system (ARMS) methods again.
Results: PIK3CA mutation rates were identified as 11.3% (12/106) by ME-Liquidchip. 10 mutations occurred in exon 9 and 2 in exon 20, including G1624A:E542K (n = 4), G1633A:E545K (n = 6) and A3140G:H1047R (n = 2). The results were further verified by ARMS methods. Among these 12 cases characterized for PIK3CA mutation, however, only 7 and 6 cases were identified by Sanger sequencing (6.6%,7/106) and pyrosequencing (5.7%,6/106), respectively.
Conclusion: Sanger sequencing and pyrosequencing are less sensitive and are not efficiently applicable to the detection of PIK3CA mutation in EC samples. Choosing between ME-Liquidchip and ARMS will depend on laboratory facilities and expertise.
Virtual slides: The virtual slide(s) for this article can be found here: http://www.diagnosticpathology.diagnomx.eu/vs/13000_2014_153.
Figures
Similar articles
-
PIK3CA mutation status in Japanese esophageal squamous cell carcinoma.J Surg Res. 2008 Apr;145(2):320-6. doi: 10.1016/j.jss.2007.03.044. Epub 2008 Feb 11. J Surg Res. 2008. PMID: 18262558
-
Frequency, characterization, and prognostic analysis of PIK3CA gene mutations in Chinese esophageal squamous cell carcinoma.Hum Pathol. 2014 Feb;45(2):352-8. doi: 10.1016/j.humpath.2013.09.011. Epub 2013 Oct 3. Hum Pathol. 2014. PMID: 24360885
-
Concordance between PIK3CA mutations in endoscopic biopsy and surgically resected specimens of esophageal squamous cell carcinoma.BMC Cancer. 2017 Jan 9;17(1):36. doi: 10.1186/s12885-016-3041-3. BMC Cancer. 2017. PMID: 28068934 Free PMC article.
-
PIK3CA gene mutations and overexpression: implications for prognostic biomarker and therapeutic target in Chinese esophageal squamous cell carcinoma.PLoS One. 2014 Jul 23;9(7):e103021. doi: 10.1371/journal.pone.0103021. eCollection 2014. PLoS One. 2014. PMID: 25054828 Free PMC article.
-
Prognostic and clinical impact of PIK3CA mutation in gastric cancer: pyrosequencing technology and literature review.BMC Cancer. 2016 Jul 7;16:400. doi: 10.1186/s12885-016-2422-y. BMC Cancer. 2016. PMID: 27388016 Free PMC article. Review.
Cited by
-
Identification of New Candidate Genes and Chemicals Related to Esophageal Cancer Using a Hybrid Interaction Network of Chemicals and Proteins.PLoS One. 2015 Jun 9;10(6):e0129474. doi: 10.1371/journal.pone.0129474. eCollection 2015. PLoS One. 2015. PMID: 26058041 Free PMC article.
-
Evolutionary history of phosphatidylinositol- 3-kinases: ancestral origin in eukaryotes and complex duplication patterns.BMC Evol Biol. 2015 Oct 19;15:226. doi: 10.1186/s12862-015-0498-7. BMC Evol Biol. 2015. PMID: 26482564 Free PMC article.
-
miR-486 functions as a tumor suppressor in esophageal cancer by targeting CDK4/BCAS2.Oncol Rep. 2018 Jan;39(1):71-80. doi: 10.3892/or.2017.6064. Epub 2017 Nov 1. Oncol Rep. 2018. PMID: 29115564 Free PMC article.
-
Anti-tumor efficacy of theliatinib in esophageal cancer patient-derived xenografts models with epidermal growth factor receptor (EGFR) overexpression and gene amplification.Oncotarget. 2017 Apr 19;8(31):50832-50844. doi: 10.18632/oncotarget.17243. eCollection 2017 Aug 1. Oncotarget. 2017. PMID: 28881608 Free PMC article.
References
-
- Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous